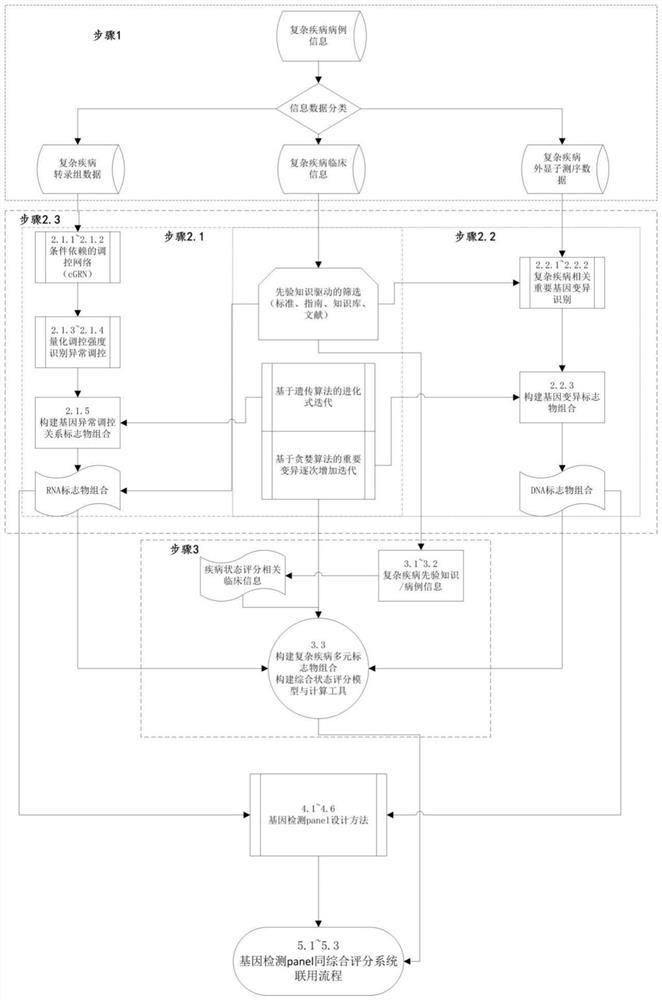
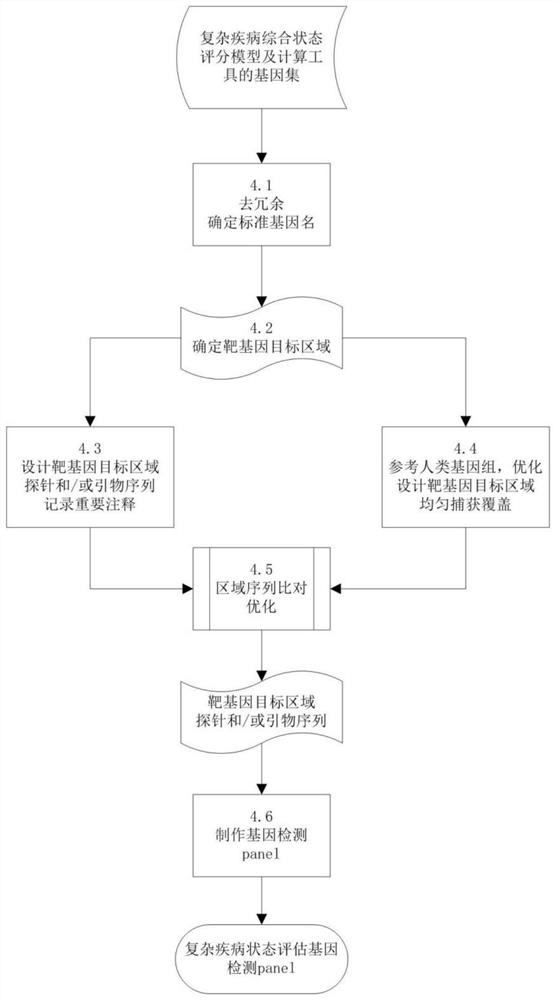
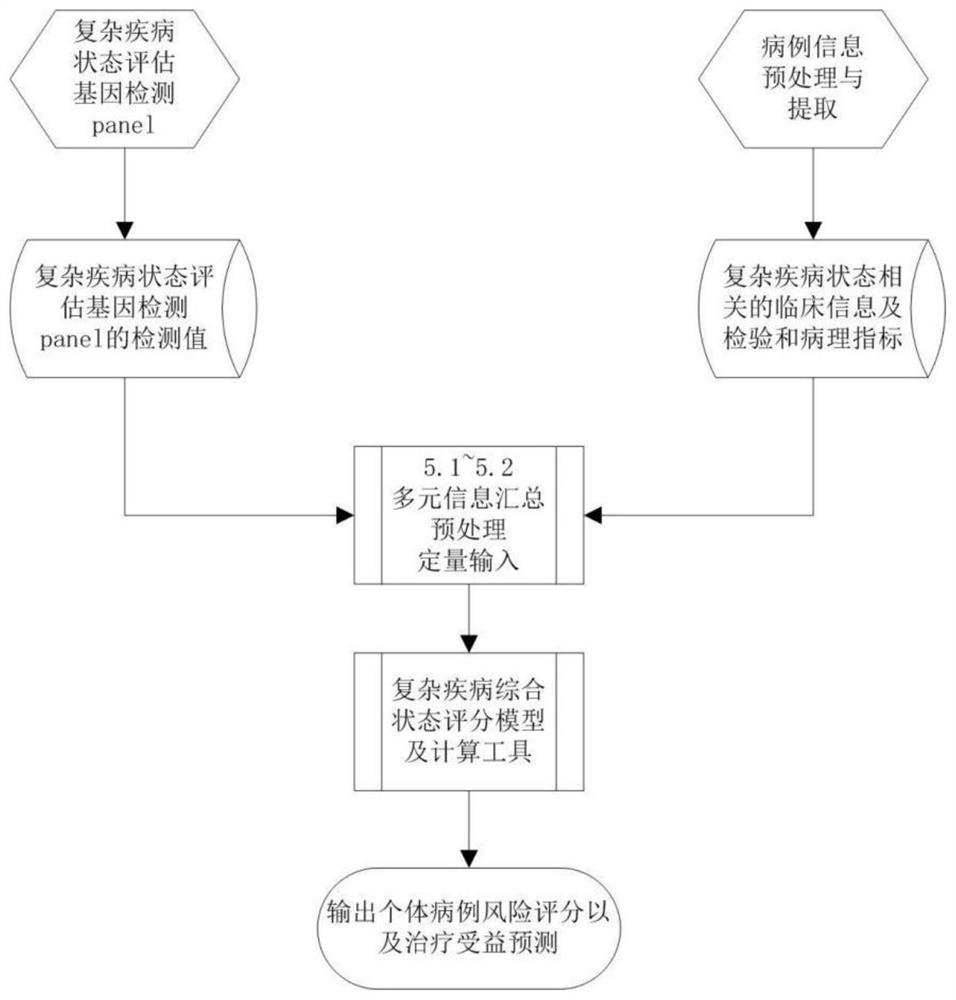
Complex disease state evaluation method based on high-throughput sequencing data and clinical phenotype construction and application
A disease state, complex technology, applied in the field of genetic testing and bioinformatics, can solve problems such as limited guiding significance, single omics, and limited functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0250] Example 1 The present invention is applied to the whole process of colorectal tumor status evaluation model construction and panel design. The present invention will be further described in detail in conjunction with specific examples. It should be understood that the following examples are only used to illustrate the present invention and not to limit the present invention. the scope of the invention. The specific steps are as follows:
[0251] S1.1 Acquisition and arrangement of colorectal tumor sequencing data and clinical phenotype information
[0252] The mRNA data and clinical data of TCGA-CRC were downloaded from the UCSC xena database. 380 orthotopic tumor samples and 51 paracancerous samples were selected. Expression levels of mRNA data were quantified in TPM. When the value of TPM is less than 1, it is regarded as a missing value. For a gene, if the number of missing values is greater than 20% of the sample size, the gene is removed. The remaining missi...
Embodiment 2
[0284] Example 2 The present invention is applied to the whole process of constructing pancreatic ductal carcinoma status evaluation model and panel design. The present invention will be further described in detail in conjunction with specific examples. It should be understood that the following examples are only used to illustrate the present invention and not to limit the present invention. the scope of the invention. Specific steps are as follows:
[0285] S2.1 Acquisition and arrangement of pancreatic ductal carcinoma sequencing data and clinical phenotype information
[0286] S2.1.1 Independently obtained the sequencing data (exome sequencing and RNA-Seq) and clinical phenotype information (including age, gender, pathological grade, surgical status R0-R2, PDX modeling status) of 71 clinical cases of pancreatic ductal carcinoma , survival including OS and DFS); PDX models were successfully established in 39 of them, and on this basis, the standard drug efficacy data of two ...
Embodiment 3
[0310] The present invention is applied to the mining of pan-tumor prognostic markers. The present invention will be further described in detail in conjunction with specific examples. It should be understood that the following examples are only used to illustrate the present invention and not to limit the scope of the present invention. Specific steps are as follows:
[0311] S3.1 Pan-tumor sequencing and collection of clinical phenotype datasets
[0312] The mRNA data and clinical data of TCGA pan-cancer were downloaded from UCSC xena. The mRNA data is derived from the data generated by the TOIL RNA-seq analysis pipeline, and the expression level of the gene is quantified by TPM. For each cancer type, tumor in situ samples and paracancerous samples were selected. Cancer types with a paired number of in situ tumor samples and paracancerous samples greater than or equal to 20 were selected for abnormal regulation analysis, and finally 14 cancer types were selected. For the m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com