Method and kit for non-specifically amplifying natural short-fragment nucleic acid
A non-specific, short-segment technology, applied in the field of molecular biology, which can solve the problems that cannot replace natural short-segment nucleic acid test samples, standards, quality control substances and reference substances, and it is difficult to achieve the size and variability of natural short-segment nucleic acids. To improve detection sensitivity and accuracy, reduce preference, and increase content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: non-specific amplification of plasma cell-free DNA according to the method of the present invention
[0045] 1. Preparation of plasma cell-free DNA
[0046] Cell-free DNA in the peripheral blood of pregnant women was extracted using a plasma cell-free DNA extraction kit (Hangzhou Berry Hekang Gene Diagnostics Technology Co., Ltd., Cat. No. R0011), and 4 ng of plasma DNA was obtained.
[0047] 2. End repair and add A
[0048] Use the NEBNext End Repair / Add dA Tail Module Kit (NEB, E7442S) to prepare the reaction system shown in Table 1 for end repair and 3' end A treatment at the same time. The reaction program is: 37°C, 20 minutes; 72°C, 20 minutes; store at 4°C. No purification was required after the reaction, and the next experiment was carried out directly.
[0049] Table 1
[0050]
[0051]
[0052] 3. Connect the connector
[0053] (1) Preparation of joints
[0054] Synthesize the sequence shown in Table 2, wherein SEQ ID NO: 1 has a deoxy...
Embodiment 2
[0085] Embodiment 2: non-specific amplification of tumor plasma cell-free DNA according to the method of the present invention
[0086] The steps of this example are the same as Example 1, the only difference is that the original DNA samples are different. Specifically, agMAXCell-Free DNA Isolation Kit (Thermo Fisher, Cat. No. A29319) was used to extract free DNA from tumor plasma to obtain 6 ng of free DNA. After non-specific amplification, its yield was 132ng, which was 22 times higher than the original plasma DNA concentration.
Embodiment 3
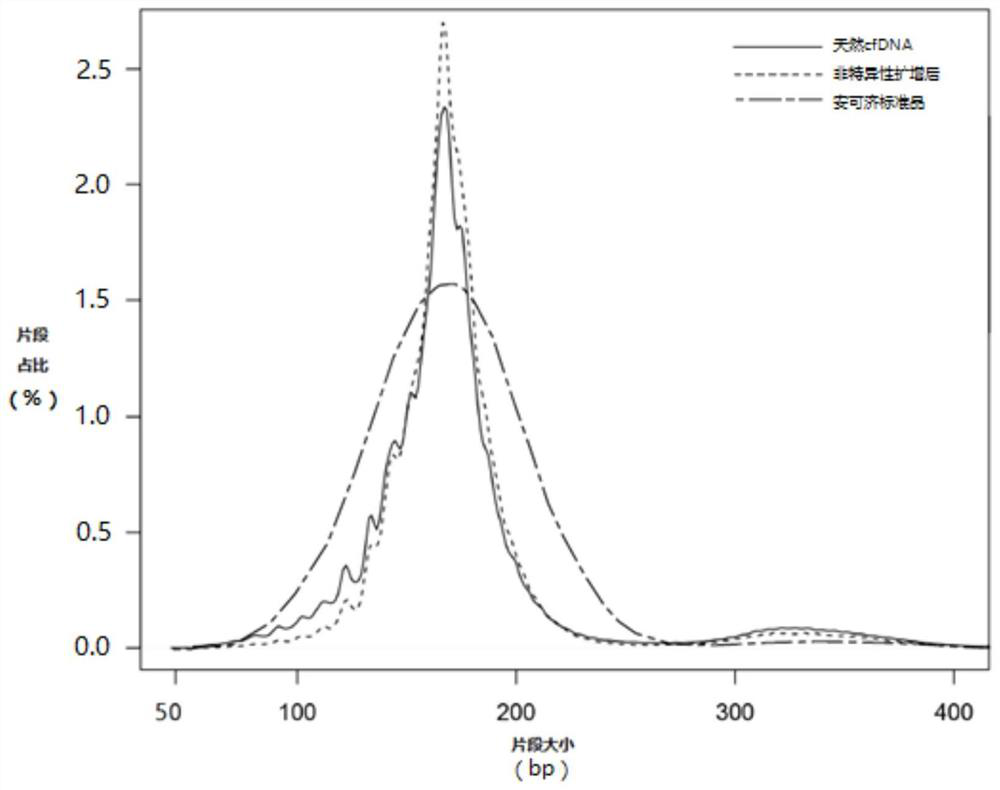
[0087] Example 3: Quality assessment of non-specifically amplified natural short fragment nucleic acids
[0088] Get 4ng of natural plasma cell-free DNA and the non-specific amplification product prepared according to the method of Example 1 respectively, according to the manufacturer's instructions, use the fetal chromosomal aneuploidy (T13 / T18 / T21) detection kit (Beijing Berry and Kang Company, Cat. No. R1000) to construct a sequencing library. Then, the Nextseq CN500 sequencer was used for sequencing, and the sequencing type was 36SE, and 5M data were measured for each sample. The quality of library sequencing data is shown in Table 8. Among them, Total Rds is the total number of sequences measured, Uniq% is the percentage of DNA sequences uniquely aligned to the human genome (hg19) in Total Rds, Redundancy% is the percentage of redundant reads in Total Rds, and Unimap_GC% is The GC percentage of the DNA sequence uniquely aligned to the human genome (hg19).
[0089] Table ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com