RNA aptamer specifically bound with CD44-hyaluronic acid binding domain protein as well as screening method and application thereof
A CD44-, hyaluronic acid technology, applied in the field of biomedicine, can solve the problems of high cost, long cycle, large labor, etc., and achieve the effects of low cost, easy storage and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
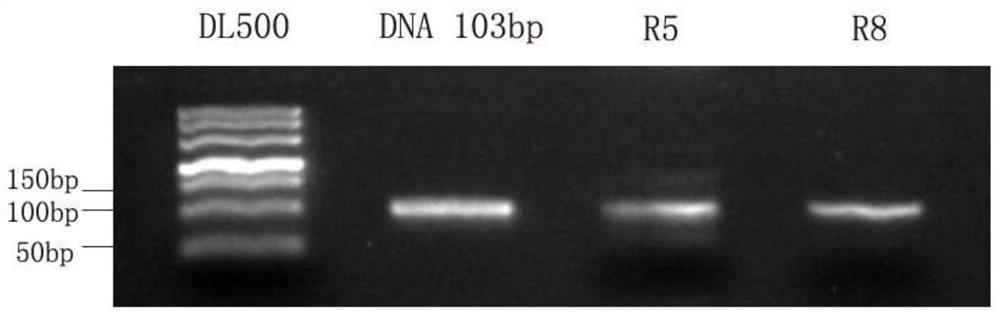
[0046] Example 1 Screening of nucleic acid aptamers.
[0047] 1. Preparation of RNA library
[0048] (1) Using the following sequences as templates and primers, a large amount of DNA library was synthesized by PCR reaction.
[0050] 5'-CACTAATACGACTCACTATAGGGAGAGAACAATGACCTN(40)GAGTGCATTGCATCACGTCAGTAG-3', wherein, N is any one of A, T, C, G;
[0051] DNA library amplification forward primer (34bp):
[0052] 5'-CACTAATACGACTCACTATAGGGAGAGAACAATG-3'
[0053] DNA library amplification reverse primer (24bp):
[0054] 5'-CTACTGACGTGATGCAATGCACTC-3'
[0055] (2) In vitro transcription of RNA library.
[0056] Prepare 100 μL in vitro transcription system according to the following conditions:
[0057]
[0058] The composition of the 10×transcription reaction buffer is 400mM Tris-Cl, 80mM MgCl2, and 20mM spermidine.
[0059] Place in a 37°C water bath and react for 8 hours.
[0060] Put the reaction system in a water bath at 90°C for 2 minut...
Embodiment 2
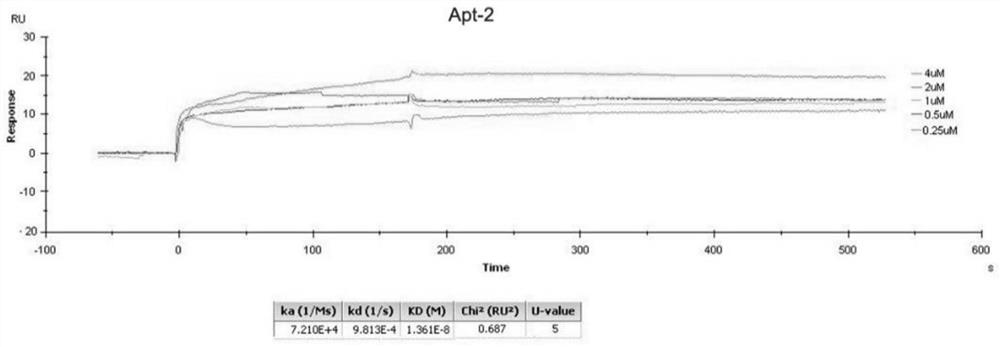
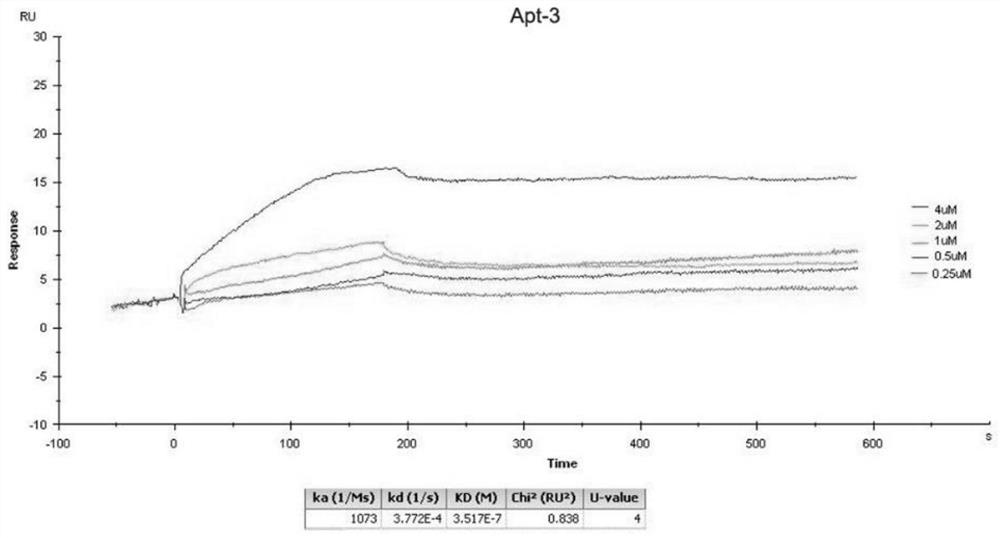
[0126] Example 2 Determining the affinity between RNA and target protein.
[0127] The coupling conditions were optimized according to the operation guide of BiacoreX100 control soft, and the CD44-HABD was diluted to 10 μg / ml and then coupled to the CM5 chip. The coupling level is 500RU, and the actual coupling amount is 518.9RU.
[0128] Detection K D
[0129] Dilute the RNA series samples and negative control (RNA library transcribed from the starting DNA library) with pH 7.4 loading buffer, and dilute a series of concentrations to 0 μM, 0.25 μM, 0.5 μM, 1.0 μM, 2.0 μM, 4.0 μM. Set the injection time to 180s, the dissociation time to 6min, and use 0.1% SDS as the regeneration buffer. Follow the operation guide of BiacoreX100control soft to carry out the computer test.
[0130] Detection of the K of each Apt by SPR D value, as shown in the table below.
[0131] Table 3. SPR results
[0132] sample name Apt-1 Apt-2 Apt-3 S2 S1 negative control D...
Embodiment 3
[0134] Example 3 Determines the specificity of RNA binding.
[0135] The CD34 protein was used as a control to coat the CM chip and perform SPR detection. The results showed that Apt-2, Apt3, and S2 had no binding and no response to the CD34 protein, indicating that the screened RNA aptamers had good specificity.
[0136] The single-stranded oligonucleotide (RNA) capable of specifically binding to human CD44-HABD was screened by optimized in vitro-SELEX technology, and the affinity coefficient of the optimal sequence binding to CD44-HABD was 13.6nM. The nucleic acid aptamer has similar functions to antibodies, but has more advantages than antibodies, such as: no immunogenicity; large-scale chemical synthesis, low cost; good stability, easy storage, etc., therefore, these nucleic acid aptamers body has broad application prospects.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com