Glyceride lipase mutant G28C-P206C as well as coding gene and application thereof
A G28C-P206C, lipase technology, applied in the field of enzyme engineering, can solve problems such as unfavorable industrialization, limited application scope, easy inactivation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1, the construction of glyceride mutant lipase expression vector
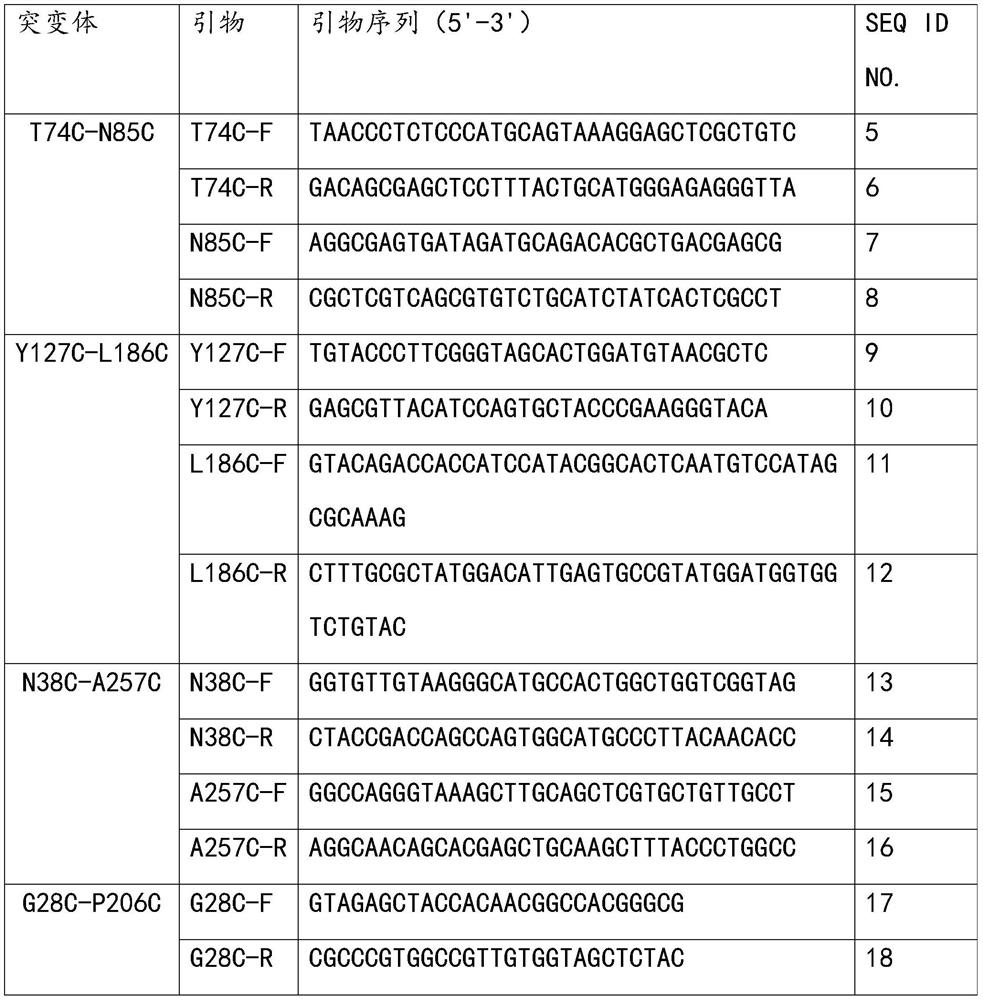
[0041] The expression vector pPICZaA-SMG1 of glyceride lipase SMG1 was constructed in the early stage of this experiment, that is, the gene of SMG1 (Genbank ID: XM_001732152.1) was inserted into the vector pPICZaA, and the restriction sites were EcoRI and SalI. Analyzing the crystal structure of lipase SMG1 (PDB ID: 3UUE), based on the experience accumulated by the inventors, and using Disulfide byDesign software to predict potential disulfide bond mutation sites, after repeated comparisons by the inventors, 5 disulfide bonds were finally left Key mutants, i.e. glyceride lipase mutant T74C-N85C, glyceride lipase mutant Y127C-L186C, glyceride lipase mutant N38C-A257C, glyceride lipase mutant G28C-P206C or glyceride lipase mutant Body T42C-F286C. Each disulfide bond mutant involves a combination of two mutation sites, that is, two amino acid residues are mutated into Cys, and the two Cys will l...
Embodiment 2
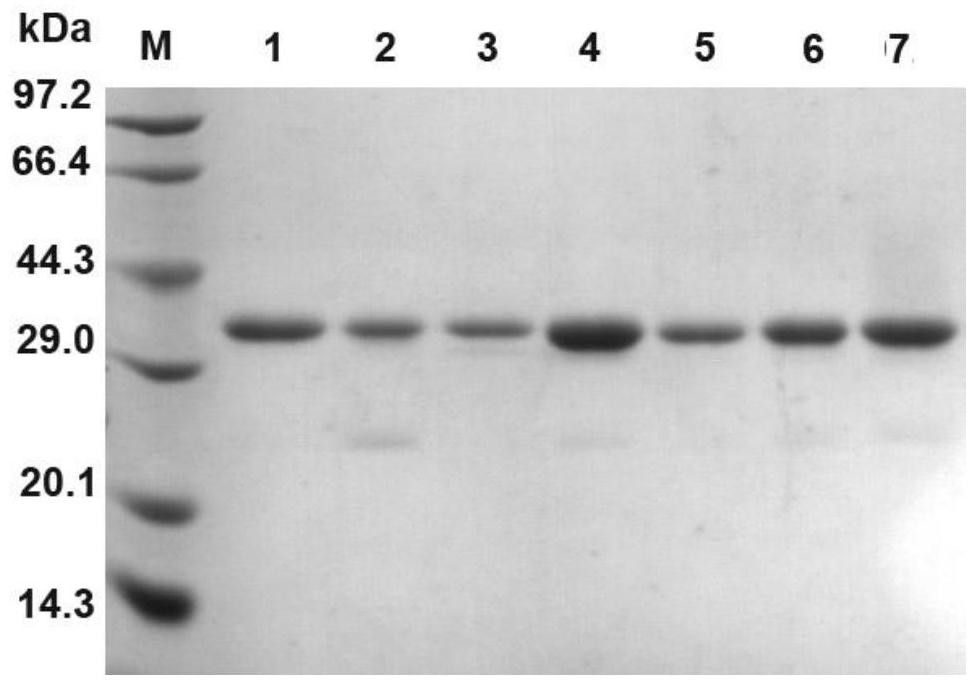
[0052] Embodiment 2: Construction, expression and purification of glyceride lipase mutant expression strain
[0053] After the positive transformants with correct sequencing were amplified overnight in LLB liquid medium, the plasmids were extracted, linearized with PmeI, purified and recovered, and transformed by electroporation with a total of 5 μg of plasmid linearized products mixed with X33 Pichia pastoris competent . Competent preparation of Pichia pastoris refers to the operation manual of Invitrogen Company. The electroporation program was set according to the parameters recommended by Bio-Rad.
[0054] Add 1 mL of 1mol / L sorbitol solution immediately after electroporation, incubate and recover the bacterial solution at 30°C for 1 hour, and spread it evenly on the YPDS+Zeocin (Zeocin concentration is 100 μg / ml) resistance plate for screening; after culturing for 72 hours, select positive Turn.
[0055] A single colony of the engineering strain was inoculated into 50 ...
Embodiment 3
[0057] Example 3: DSF Screening of Glyceride Lipase Mutants
[0058] Dilute the target protein to the same concentration of 0.3mg / ml, and dilute the dye Sypro Orange dye 100 times. Mix 20 μL of protein with 5 μL of dye, and perform protein thermal inactivation curve measurement in Bio-Rad’s real-time fluorescent quantitative PCR instrument to obtain the T of the protein. m value. The results are shown in Table 2, the T of glyceride lipase mutant G28C-P206C m It is 9.0°C higher than the wild type and 3.0°C higher than the mutant S5.
[0059] Table 2 DSF measurement results
[0060]
PUM
| Property | Measurement | Unit |
|---|---|---|
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com