Method for detecting food-borne pathogenic bacteria based on double signals of DNA Walker driven by chain substitution triggered by nucleic acid conformation
A technology of food-borne pathogenic bacteria and chain substitution, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection, etc. Easy to operate and avoid false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
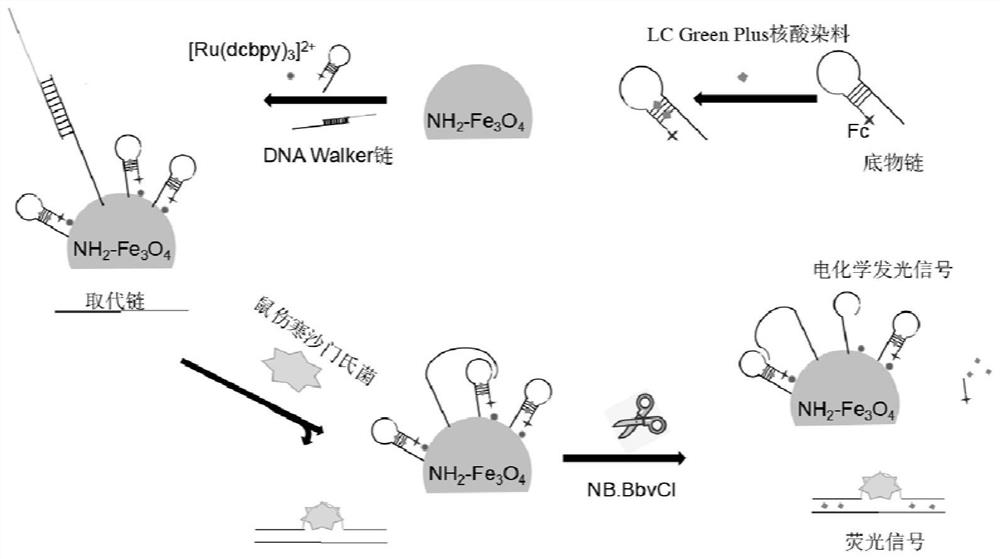
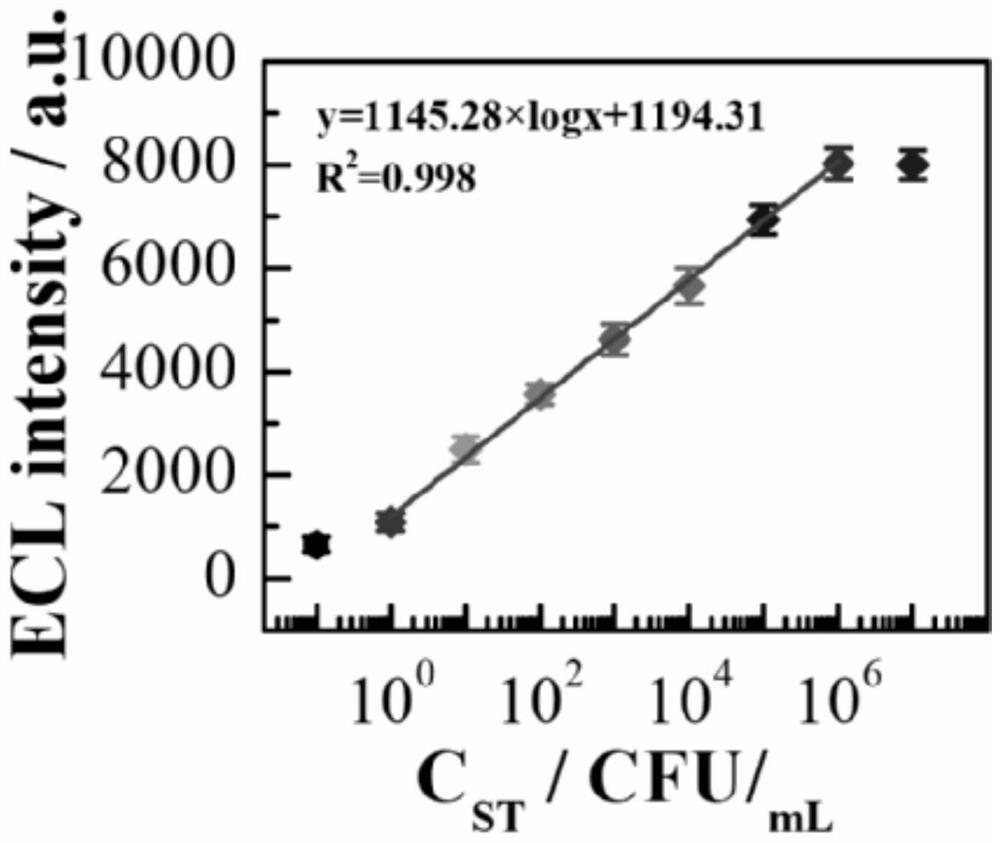
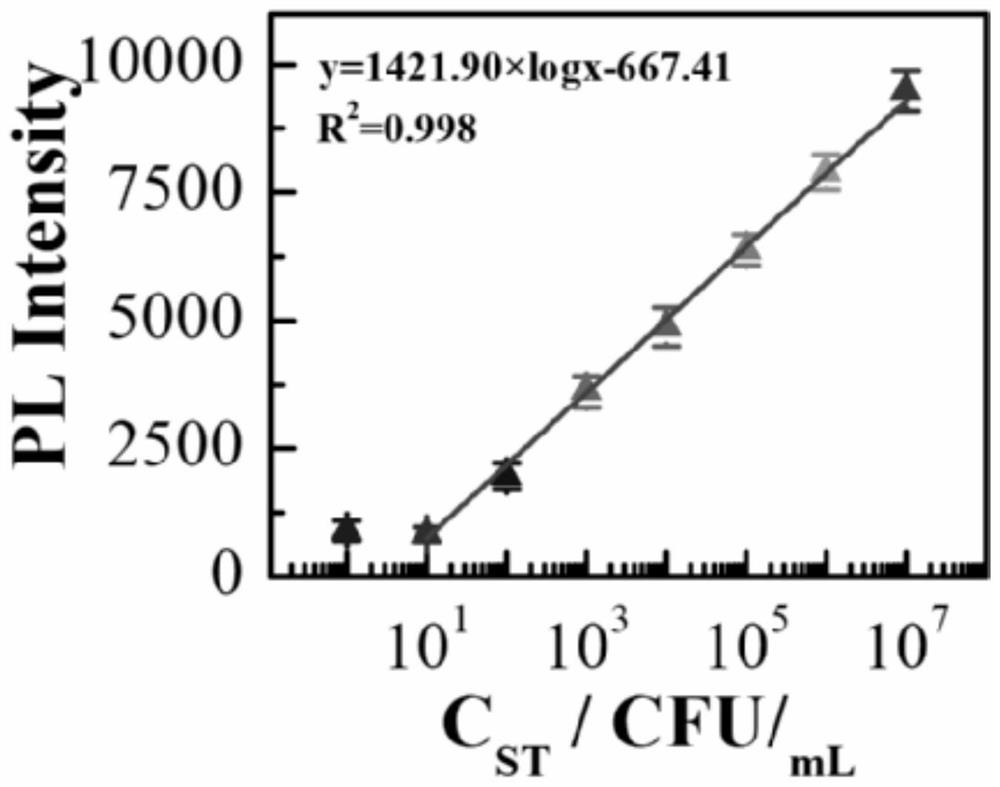
[0033] A dual-signal detection method for Salmonella typhimurium based on nucleic acid conformation-initiated strand displacement-driven DNA Walker, such as figure 1 shown, including the following steps:
[0034] (1) After mixing equal volumes of 15 μmol / L DNA Walker swing arm chain solution and 15 μmol / L protection probe solution containing the aptamer of Salmonella typhimurium to be tested, place it at 95 ℃ for 8-12 minutes to deform Slowly lower to room temperature to form a protected DNA Walker chain solution; wherein the solvents of the swing arm chain solution and the protection probe solution are oligonucleotide stock solutions; the sequence of the Salmonella typhimurium aptamer is TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG; the corresponding protection probe The sequence of the needle is: 5'-CTTCAAGGCTAACATGGTATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAGGAGGCAAGTGATCCGG-3'; the sequence of the DNA Walker swing arm strand is: 5'-NH 2 -TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGA...
Embodiment 2
[0043] With above-mentioned embodiment 1, its difference is:
[0044] (1) After mixing equal volumes of 10 μmol / L DNA Walker swing arm strand solution and 10 μmol / L protection probe solution containing the aptamer of Salmonella typhimurium to be tested, the oligonucleotide stock solution is pH=7 containing 1 mM EDTA and 10mM MgCl 2 •6H 2 30 mM Tris-HCl buffer solution of O;
[0045] (2) Add 3 µL of a nucleic acid dye capable of intercalating between double strands (LC Green Plus nucleic acid dye) into 500 µL of 8 µM hairpin substrate strand solution, shake at room temperature for 4-6 min;
[0046] (3) Mix 1 mL of 10 mg / mL aminated ferric oxide and 50 µL of glutaraldehyde, stir at room temperature for 50 min, suspend in 2 mL of oligonucleotide stock solution after magnetic cleaning, and then add fully mixed Uniformly mix 50 µL of 10 µM protected DNA Walker strand solution and 500 µL of 8 µM hairpin substrate strand mixture, react at 35 °C for 20 min, and dilute to 2 mL with ...
Embodiment 3
[0049] With above-mentioned embodiment 1, its difference is:
[0050] (1) After mixing equal volumes of 20 μmol / L DNA Walker swing arm strand solution and 20 μmol / L protection probe solution containing the aptamer of Salmonella typhimurium to be tested, the oligonucleotide stock solution is pH=9 containing 3 mM EDTA and 13 mMMgCl 2 •6H 2 50 mM Tris-HCl buffer solution of O;
[0051] (2) Add 5 µL of nucleic acid dye (LC Green Plus nucleic acid dye) capable of intercalating between double strands to 600 µL of 5 µM hairpin substrate strand solution, shake at room temperature for 4-6 min;
[0052] (3) Mix 2 mL of 5 mg / mL Fe3O4 and 100 µL of glutaraldehyde, stir at room temperature for 70 min, suspend in 5 mL of oligonucleotide stock solution after magnetic cleaning, and then add fully mixed Uniformly mix 60 µL 5 µM protected DNA Walker strand solution and 600 µL 5 µM hairpin substrate strand mixture, react at 38°C for 40 min, and dilute to 5 mL with oligonucleotide stock soluti...
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com