Mung bean InDel molecular marker detection primer group and application thereof
A technology of molecular markers and detection primers, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problem of low polymorphic information content, achieve high polymorphism, good repeatability, Rich number of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1I
[0019] Embodiment 1InDel mark development
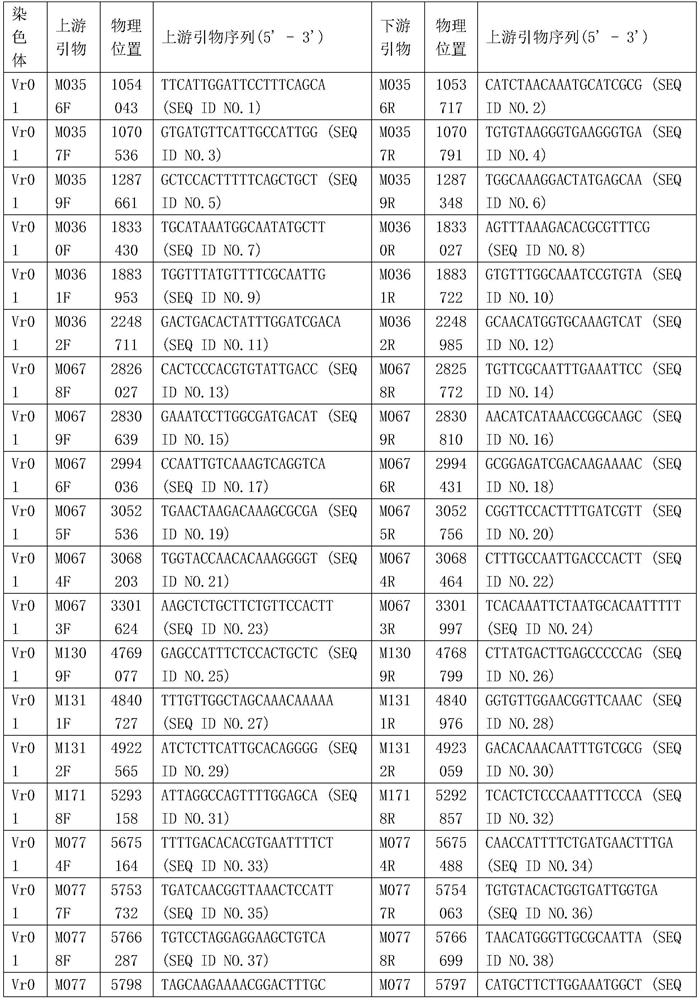
[0020] After extracting the genomic DNA of mung bean varieties Sulu 1 and V2709, using illumine SBS constructed the library and performed whole genome sequencing using illumine MiSeq sequencing. After removing adapters and filtering, paired-end sequence data of 20,526Mb and 17,944Mb were obtained for Sulu 1 and V2709, respectively. Use the workflow of mInDel software to develop InDel markers, and the expected InDel size parameters are set to 20-500bp. As a result, 1809 pairs of candidate InDel-labeled primers were generated.
[0021] The resulting InDel-marked primer sequences were compared to the mung bean reference genome sequence (GenBank assembly accession: GCA_000180895.1) using BLAST to obtain the physical positions of these primers. According to the comparison results, filter out some candidate markers according to the following four conditions: 1) There is a mismatch at the 3' end of the primer; 2) The front primer and th...
Embodiment 2
[0065] Embodiment 2 mung bean genetic diversity analysis
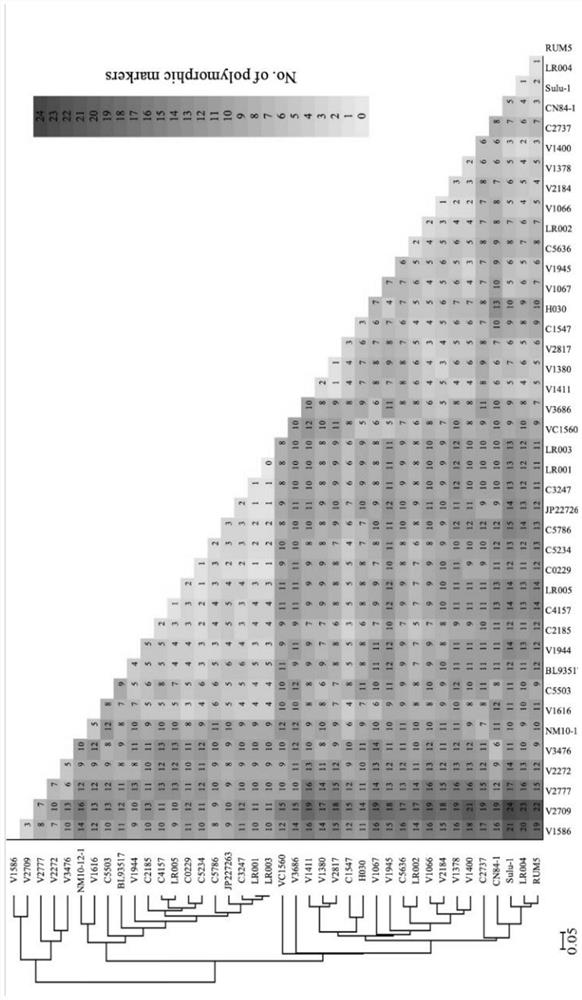
[0066] Randomly select 24 polymorphic InDel markers in Table 1, including M0082, M0110, M0115, M0130, M0183, M0189, M0310, M0366, M0550, M0616, M0793, M0826, M0841, M1160, M1229, M1252, M1308, M1309, For M1344, M1351, M1355, M1366, M1489 and M1516, a 0-1 matrix was formed for the genotyping band data of natural populations composed of 40 mung bean varieties (or germplasm resources), and the polymorphic markers among various germplasm materials were counted. number. And use powermarker to calculate the Nei’s genetic distance among various qualitative materials, and construct a phylogenetic tree according to the distance matrix, and perform Neighbor-Joining cluster analysis, and the generated results are as follows figure 2 As shown, 40 mung bean materials can be divided into two groups.
Embodiment 3
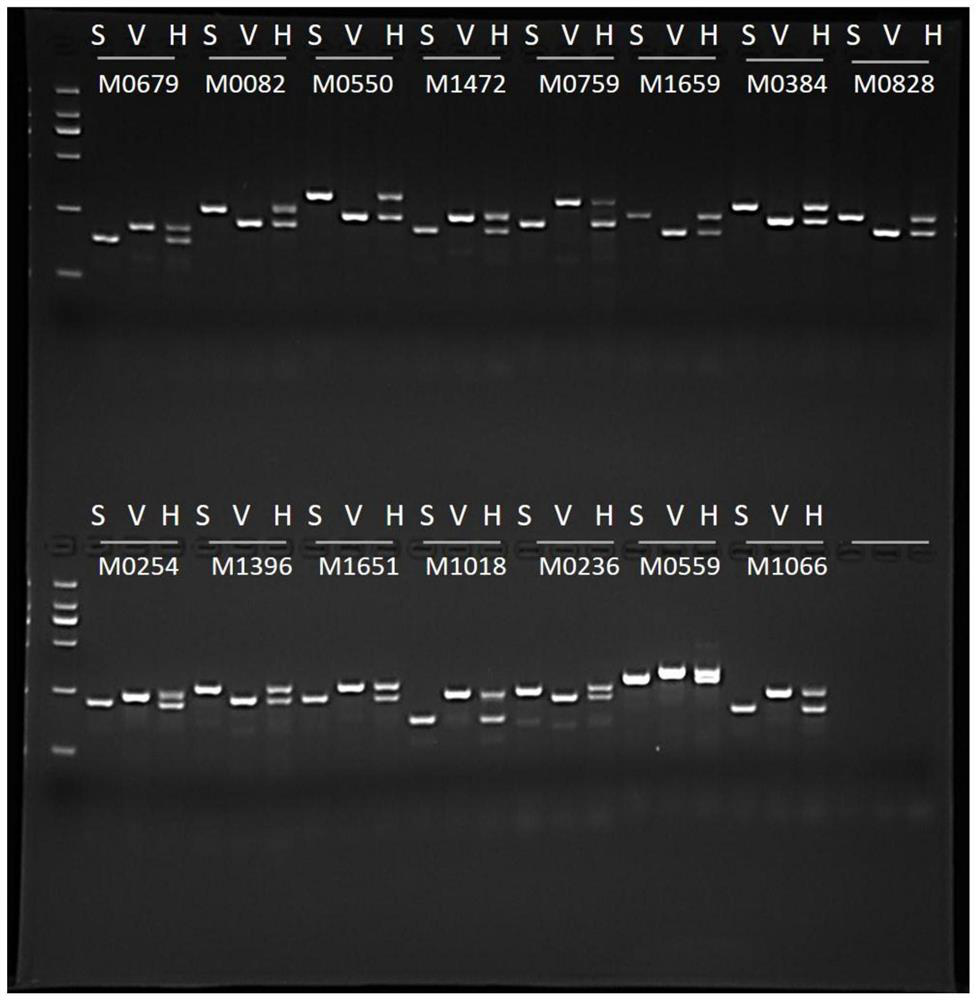
[0067] Example 3 Authenticity Identification of Mung Bean Hybrid Progeny
[0068] The mung bean variety Sulu No. 1 was used as the female parent and V2709 was used as the male parent for hybridization. The obtained seeds are planted as a single plant after maturity. Take the leaves of each hybrid plant to be tested, and use the CTAB method to extract DNA. Using the DNA of each hybrid plant to be tested and the DNA of the two parents as templates, PCR amplification was performed with InDel-marked M0679 primers. The obtained PCR products were subjected to electrophoresis in 3% agarose gel. If the hybrid plant to be tested has two bands, one of which is the same size as Sulu 1 and the other is the same size as V2709, then the plant is a true hybrid progeny. If the hybrid plant to be tested only gets one band in the same phase as Sulu No. 1, it is a false hybrid progeny.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com