Primer and probe for identifying brucella vaccine strain A19 and wild strain, and application thereof
A technology of Brucella and wild strains, applied in the direction of microorganisms, recombinant DNA technology, and methods based on microorganisms, can solve the problems of high cost of detection and typing, low detection cost, etc., and achieve fast detection speed, high sensitivity, The effect of improving correctness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Design and screening of embodiment 1 primers and probes
[0032] After screening a large number of primers and probes designed, it was found that the primer pair F, R, and probe P fluorescent method had the best effect in distinguishing Brucella A19 (S19) vaccine strains from wild strains. As follows.
[0033] Primer F: 5'-CATCGATGTGCCGTTCATCATG-3' (SEQ ID NO: 1),
[0034] Primer R: 5'-CACATCTTCGCCGACATAGC-3' (SEQ ID NO: 2),
[0035] Probe P: 5'-ATGCGACGACGCTGACGCAAGCC-3' (SEQ ID NO: 3).
[0036]A fluorescent group FAM is labeled at the 5' end of the probe P, and a quencher group BHQ1 is labeled at its 3' end.
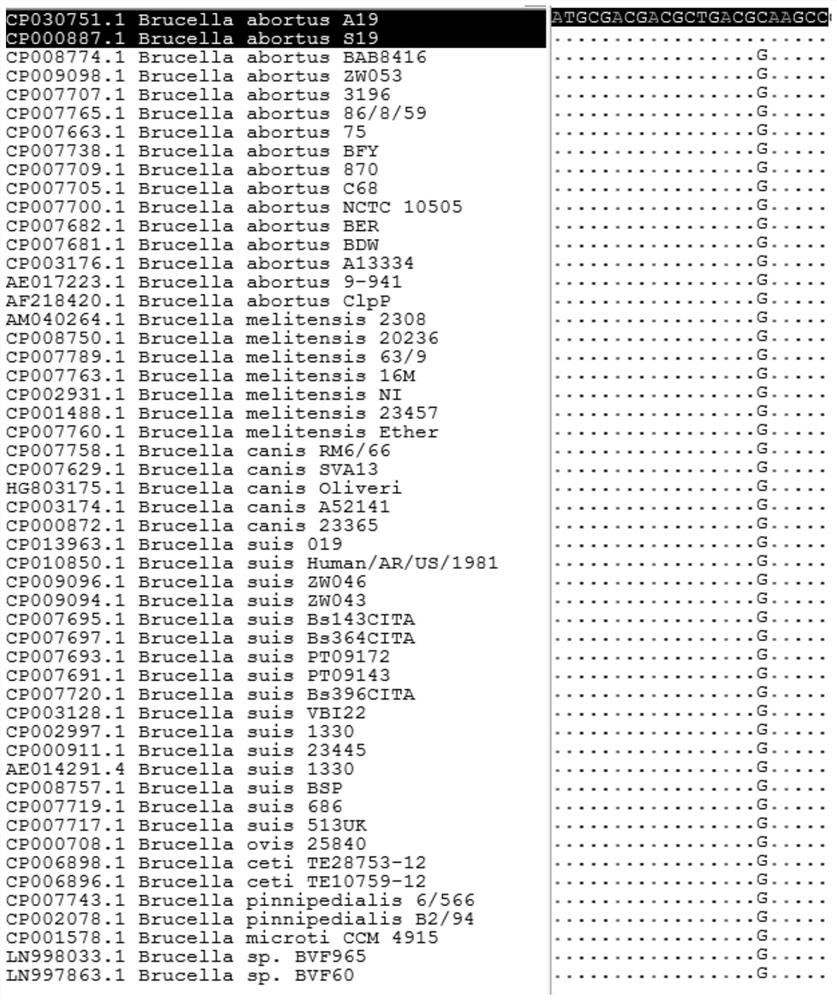
[0037] like figure 1 As shown, at the probe P position, there is only one SNP site (C18G) between the Brucella A19 (S19) vaccine strain and the wild strain.
Embodiment 2
[0038] The preparation of embodiment 2 standard samples, fluorescent PCR amplification and melting curve analysis
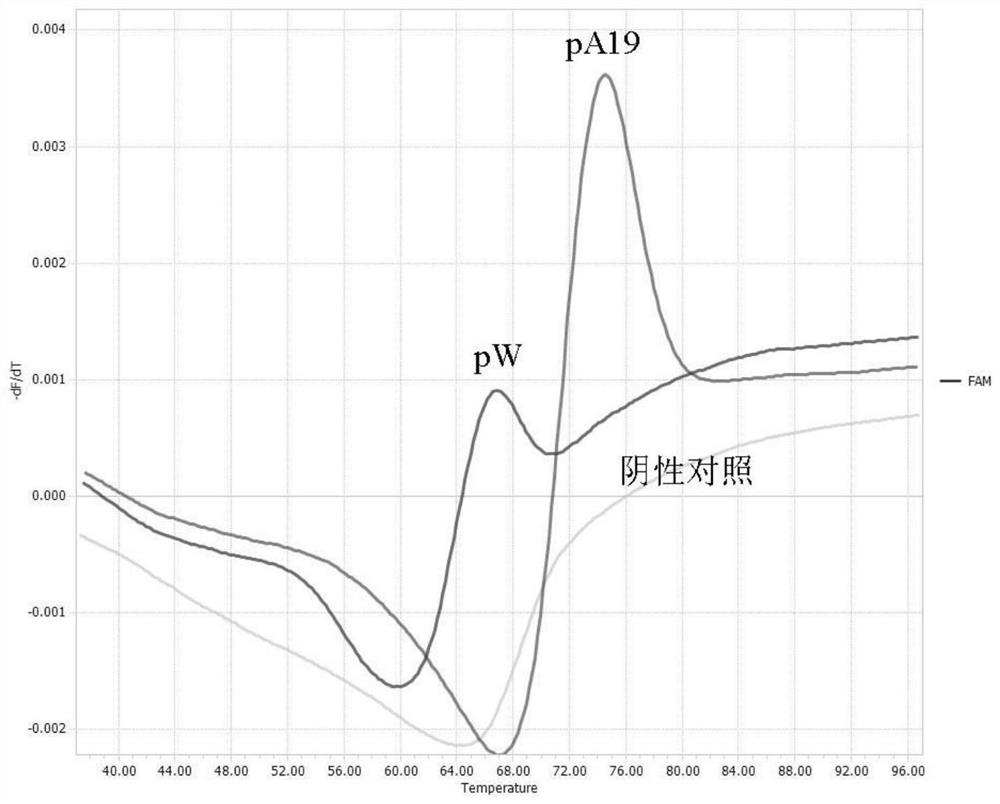
[0039] 1. Preparation of standard samples
[0040] In order to verify the feasibility and reliability of the method of the present invention, a standard positive sample was prepared to provide a positive control for subsequent method establishment, sensitivity, and specificity tests.
[0041] according to figure 1 Probe sequence comparison analysis, the corresponding Brucella A19 (S19) vaccine strain sequence of probe P is one group, and other wild strains are another group.
[0042] The genome nucleic acid of the Brucella A19 vaccine and wild strain representative strain 544A used in the present invention is donated by China Veterinary Drug Control Institute. Using the nucleic acid of the A19 (S19) vaccine and the genomic nucleic acid of the field strain 544A as templates, the following primer pairs were used:
[0043] CF: 5'-GTTCTGTCGGTTGCGGTTCA-3' (SEQ ID N...
Embodiment 3
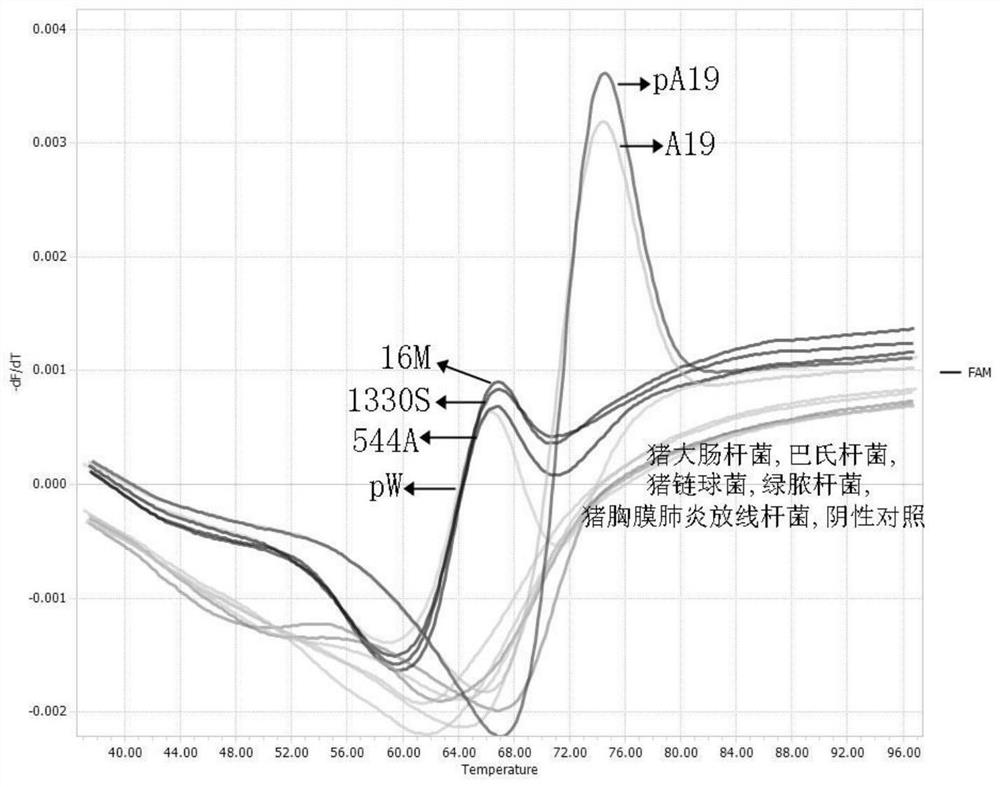
[0055] Embodiment 3 specificity experiment
[0056] The genome nucleic acid of Brucella vaccine A19 strain, Brucella bovis 544A strain, Brucella suis 1330S strain, Brucella ovis 16M strain donated by the China Veterinary Drug Administration and other common samples were respectively selected. Pathogens of common bacterial diseases in pigs isolated and preserved in the laboratory: E. coli, Pasteurella, Streptococcus suis, Pseudomonas aeruginosa, Actinobacillus pleuropneumoniae ( Actinobacillus pleuropneumoniae).
[0057] Utilize DNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.) to extract the above-mentioned bacterial nucleic acid, and use the extracted nucleic acid and water (negative control) as PCR templates respectively, with the PCR amplification reaction in the above-mentioned embodiment 2 and The melting curve analysis method was used for analysis, and compared with the positive standard samples pA19 and pW, the peak shape of the melting curve was ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com