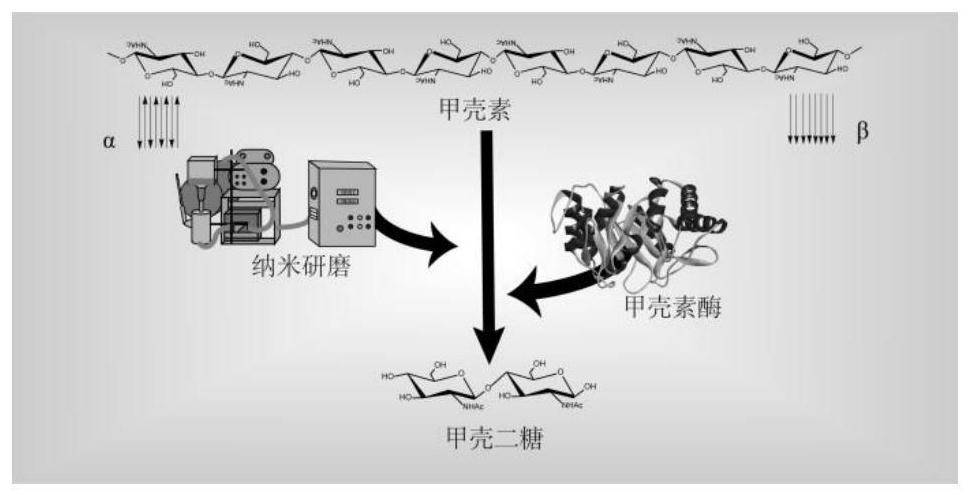
Method for preparing chitooligosaccharide
A technology of chitosan oligosaccharide and chitin, which is applied in the field of nanomaterial preparation and chitosan oligosaccharide preparation, can solve the problems of low yield, pollute the environment, use concentrated acid, etc., and achieves low crystallinity, high enzymatic hydrolysis conversion rate, and easy hydrolysis. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1 chitinase gene SbChiAJ143 clone
[0035] Streptomyces bacillaris CGMCC 4.1584 genome was used as a template for PCR amplification. The PCR reaction conditions were: pre-denaturation at 95°C for 10 minutes, denaturation at 98°C for 10 seconds, annealing at 58°C for 30 seconds, extension at 68°C for 3 minutes, 30 cycles of reaction, and extension at 68°C 10min.
[0036] The nucleotide sequences of the specific primers used for PCR amplification are as follows:
[0037] Upstream primer: 5'-GGCCGCAAGCTTCGGCAGGCTGCGGAC-3';
[0038] Downstream primer: 5'-CAAATGGGTCGCGGATCCATGGTGCAACGCTCCG-3'.
[0039] The target gene was connected with the pET-28a(+) cloning vector using seamless connection technology, and the connection product was transferred into E.coliDH5α competent cells, and the sequencing was successful after positive verification.
Embodiment 2
[0040] Embodiment 2 recombinant protein expression and purification
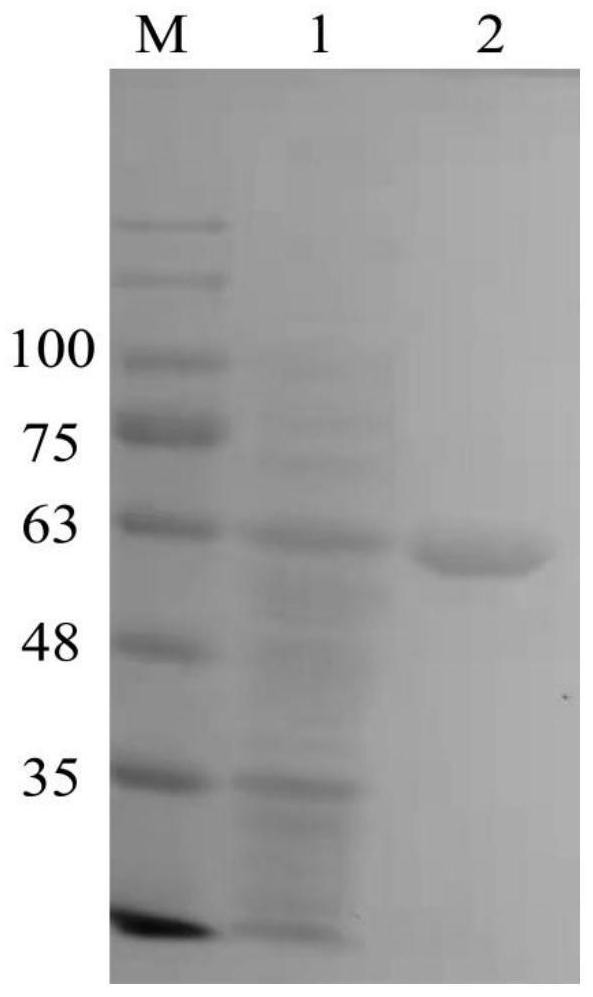
[0041] The recombinant plasmid was transferred into the expression strain E.coliBL21(DE3), cultured at 20°C, 220rpm for 48 hours, and the cells were collected (centrifuged at 8000g for 10 minutes at 4°C). The bacteria were resuspended in 50mM Tirs-HCl buffer solution with pH 7.4, sonicated for 50min and then centrifuged at 12000g for 20min, and the supernatant was the crude enzyme solution. Purify the crude enzyme solution by affinity chromatography with a Ni-NTA column, use 10mM imidazole solution (500mM NaCl, 50mM Tris-HCl) to equilibrate the column, and then use 20mM, 40mM imidazole solution (500mM NaCl, 50mM Tris-HCl) to elute the binding Weak foreign protein, 60mM imidazole solution elutes to obtain the target protein, and the obtained solution is subjected to SDS-PAGE detection, such as figure 2 As shown, the molecular weight of the obtained target protein is about 59.2KD, which is equivalent to chit...
Embodiment 3
[0042] Embodiment 3 Recombinant chitinase optimum pH and stability determination
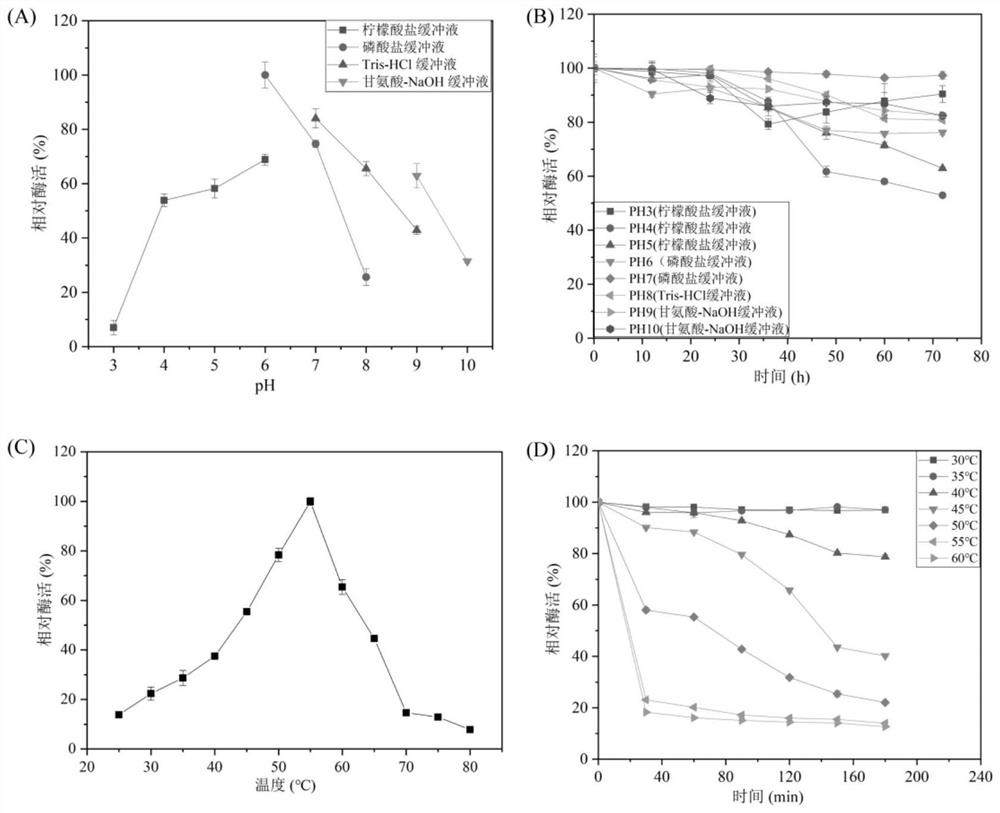
[0043] Method for determining the optimum pH value: 10 μL of enzyme solution (prepared in Example 2) was added to 1% colloidal chitin substrate prepared by 190 μL of different pH buffers, respectively: citric acid buffer (pH 3.0- 6.0), phosphate buffer (pH 6.0-8.0), Tris-HCl buffer (pH 7.0-9.0) and glycine-NaOH buffer (pH 9.0-10.0), react at 55°C for 30min. The DNS method was used to measure the activity of the pure enzyme, and the results were as follows: image 3 As shown in A, it can be seen that the optimum reaction pH is pH 6.0 (phosphate buffer).
[0044] The pH stability test method is: add pure enzyme (prepared in Example 2) to the above pH buffer solution, place it at 25°C for 0, 12, 24, 36, 48, 60, and 72 hours, take out 10 μL, add colloidal chitin 1% substrate, determine the residual enzyme activity, the results are as follows image 3 As shown in B, it can be seen that within the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| crystallinity index | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com