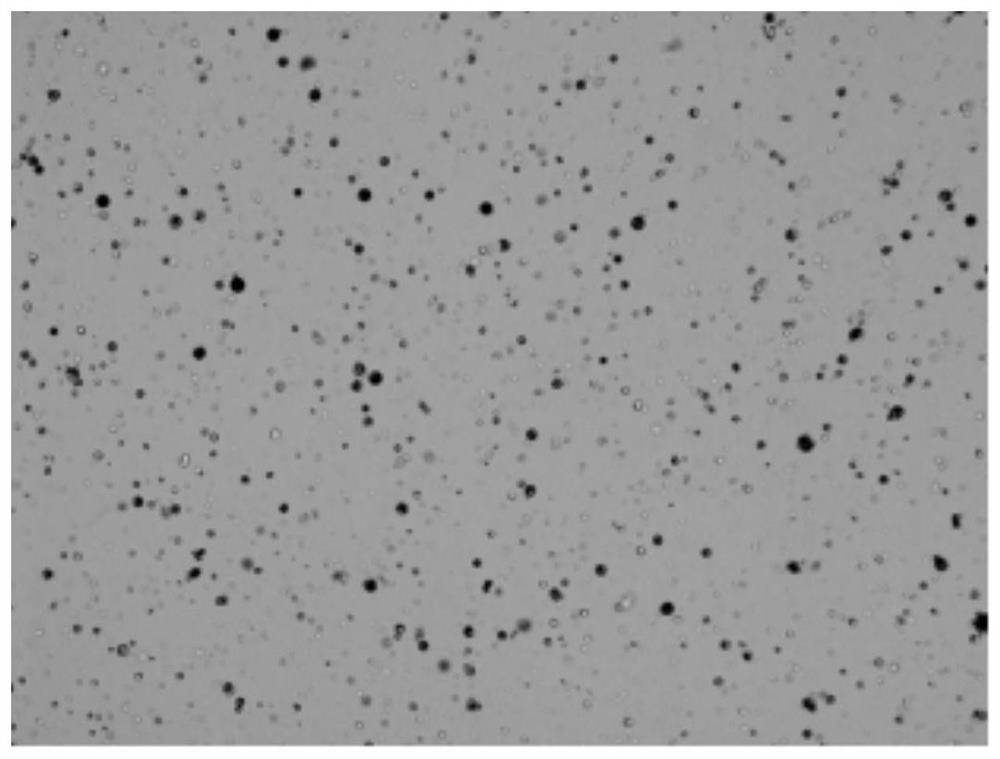
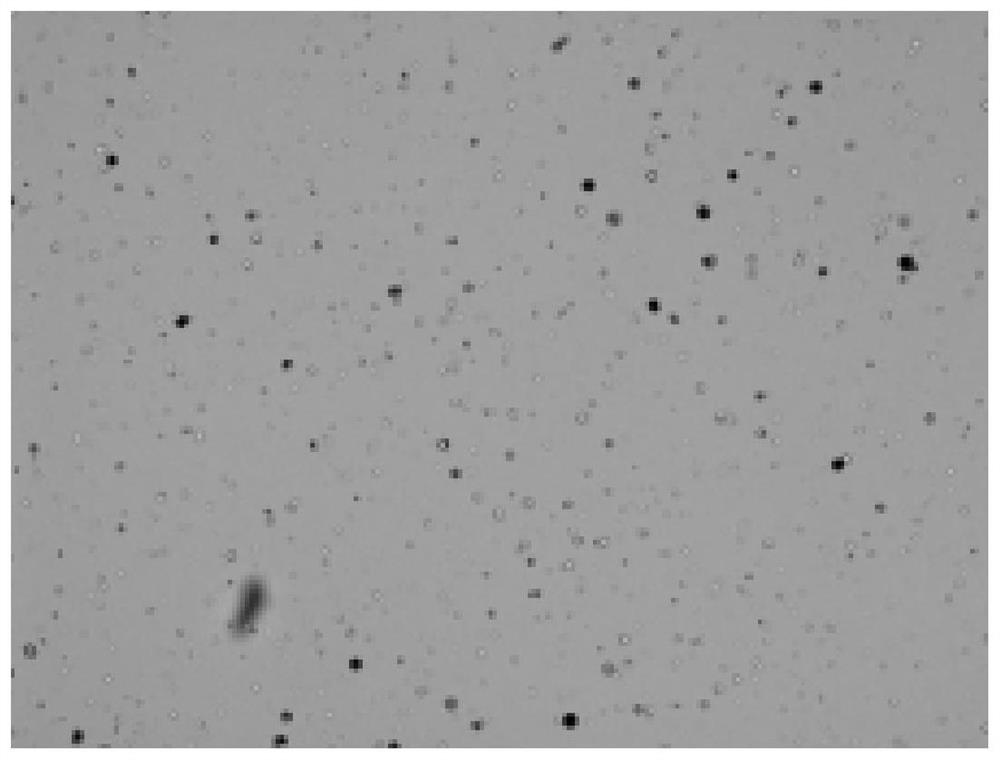
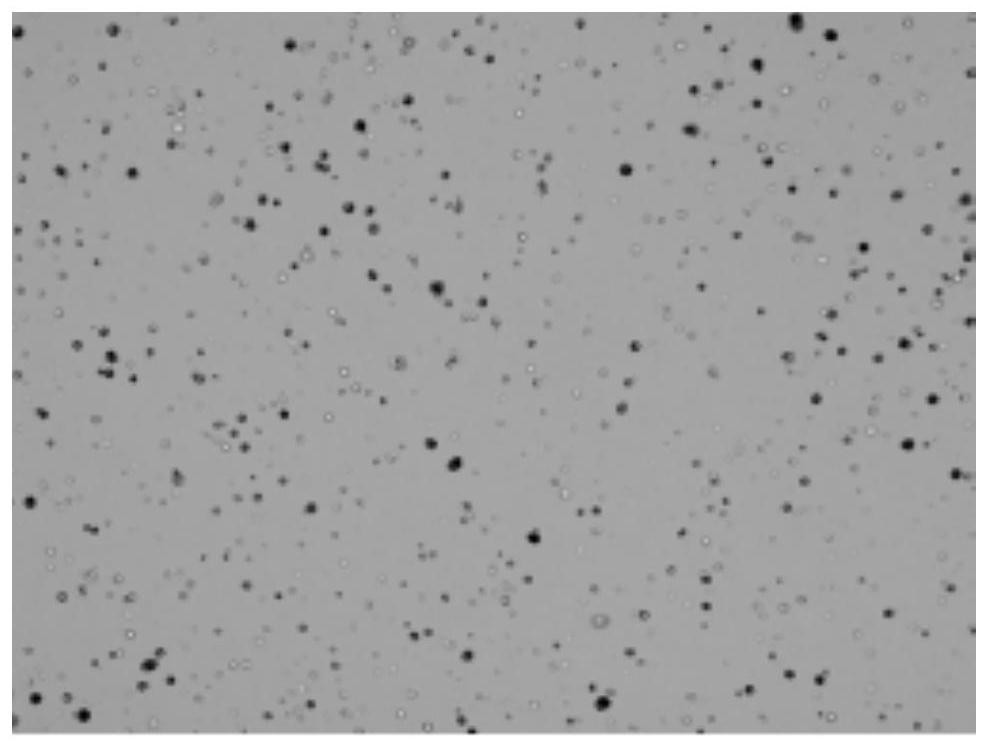
Single cell separation method of Aurelia coerulea polyps
A single-cell separation and sea-moon jellyfish technology is applied in the field of single-cell separation of sea-moon jellyfish polyps. The effect of simple separation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] A. Preparation of artificial seawater
[0035] Preparation of artificial seawater: According to Table 1, artificial seawater with a theoretical salinity of 2.8 to 3.0 was prepared as needed, and stirred while adding salt until it was completely dissolved.
[0036] When used in the laboratory, it is preferable to prepare 1L dosage: prepare 1L of distilled water, weigh 30g of sea salt and pour it into the water, add salt while stirring until it is completely dissolved, measure the salinity with a salinity meter, and control the reading between 2.8 and 3.0.
[0037] Table 1 Preparation table of artificial seawater and distilled water
[0038]
[0039] Sterilization of artificial seawater: Put the prepared artificial seawater and distilled water into 500mL Erlenmeyer flasks, each bottle contains about 300mL, wrap the mouth of the bottle with tin foil, 120°C, 5min. After the sterilization is finished, measure the salinity again with a salinity meter, and adjust the salin...
Embodiment 2
[0052] In this embodiment 2, the only difference from embodiment 1 is that the components of the dissociating enzyme solution are different, and other methods are the same as in embodiment 1. In this example, dissociation enzyme solution 2 is selected for tissue digestion, and the preparation method is as follows: take 1 mL of dissociation solution, add 1 mg of collagenase IV, and mix well with 20 U of papain.
Embodiment 3
[0054] In this embodiment 3, the only difference from embodiment 1 is that the pretreatment method of the polyp of the jellyfish is different: in this embodiment, the prepared polyp of the jellyfish is cut off the mouth, arms, tentacles and feet with a scalpel, Body dry as a sample, then cut into 0.1×0.1×0.1cm with a scalpel 3 of small pieces.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com