Method and system for screening specific BCR/TCR
A specific and sequence-based technology, applied in the fields of molecular biology and cellular immunology, can solve problems such as failure, multi-identification, and difficulty in screening human antibodies, and achieve the effects of improving efficiency, high specificity, and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
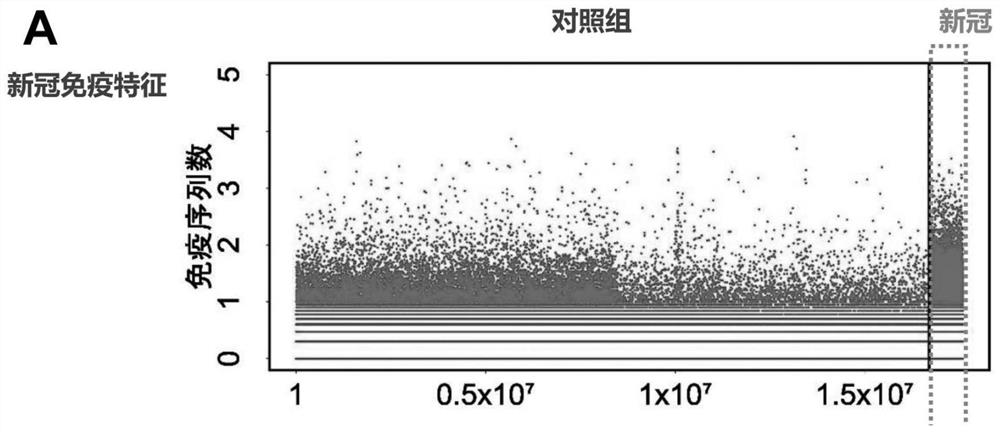
[0056] Example 1 Obtaining the Characteristic Antibody Heavy Chain Sequence Set in Peripheral Blood After Novel Coronavirus (COVID-19) Infection by High-throughput Sequencing
[0057] 1. Collect the peripheral blood of 1377 control groups (including healthy people and patients not infected with the new crown) and 94 patients infected with the new coronavirus:
[0058] (1) Use EDTA sterile blood collection tube (do not use heparin or citric acid), collect blood 0.5-1mL, flick the blood collection tube to mix the blood and anticoagulant evenly. If the collected blood is too late to be processed, it can be temporarily stored in a 4°C refrigerator (no more than 2 hours).
[0059] (2) Erect the blood collection tube, use a brand new disposable syringe, and inject 5mL TRIzol (TRIsol, RNAiso plus, SuperfecTRI all can) into the blood collection tube through the rubber stopper (just pass it, do not open the cap of the blood collection tube) . If the blood has stratified after being s...
Embodiment 2
[0125] Example 2. Screening of the most potential novel coronavirus antibody heavy chain sequence from the characteristic antibody heavy chain sequences in peripheral blood after novel coronavirus infection
[0126] 1. From the novel coronavirus characteristic antibody heavy chain sequence set, remove all antibody heavy chain sequences that appear in less than 10 new coronavirus infected samples (<10% of the number of new coronavirus infected patients). A total of 77 antibody CDR3 sequences were selected in this step.
[0127] 2. Sort the antibody heavy chain sequences screened in step 1 according to "the sum of the number of repeated occurrences of any unique antibody heavy chain CDR3 sequence in all novel coronavirus infection patient samples" from high to low, and select the top 100 Or the top 10%. Since there are few sequences selected in step 1, all of them are reserved in this step.
[0128] 3. According to the characteristics of the antigen protein, screen the antibod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com