Method for producing L-phenylglycine through biological catalysis of genetic engineering strain
A technology of genetically engineered bacteria and phenylglycine, which is applied in the field of biocatalytic production of L-phenylglycine by genetically engineered strains, and can solve the problems of low catalytic activity of sulfuric acid and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
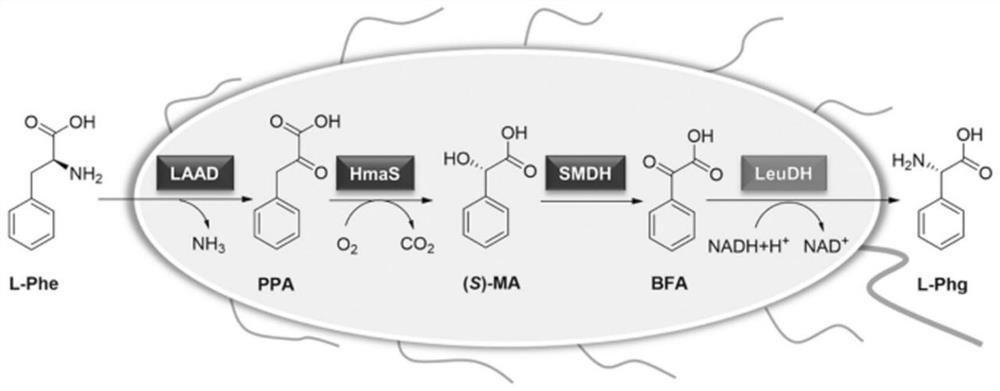
[0030] Embodiment 1 constructs recombinant escherichia coli
[0031] 1. Construction of pET-LAAD-HmaS plasmid
[0032] With the sequences shown in SEQ ID NO: 5 and SEQ ID NO: 6 as the upstream and downstream primers (Table 1), PCR amplifies the L-α-amino acid deaminase gene LAAD (SEQ ID NO: 1), and the gel recovery object band; wherein, PCR amplification conditions: 98°C for 2min, 98°C for 10s, 56°C for 30s, 72°C for 1min, 30 cycles; 72°C for 2min.
[0033] The hydroxymandelic acid synthase gene HmaS (SEQ ID NO: 2) was amplified by PCR with primers SEQ ID NO: 7 and SEQ ID NO: 8, and the target band was recovered from the gel; wherein, PCR amplification conditions: 98°C for 2min, 98°C 10s, 56°C for 30s, 72°C for 1min, cycle 30 times; 72°C for 2min.
[0034] The L-α-amino acid deaminase gene LAAD and the hydroxymandelic acid synthase gene HmaS were respectively digested with BsaI at 37°C for 2 hours, and the digested fragments were recovered by PCR purification; the enzyme dig...
Embodiment 2
[0047] Example 2 Whole Cell Biotransformation Recombinant Strain
[0048] The Escherichia coli engineering bacteria prepared in Example 1 were inoculated in 2mL LB culture solution, then inserted into 100mL TB culture solution at a ratio of 1:100 to expand the culture for 2 to 3 hours, OD 600 After reaching 0.6, add 0.5mM IPTG, induce protein expression at 22°C for 20h, and obtain bacteria by centrifugation.
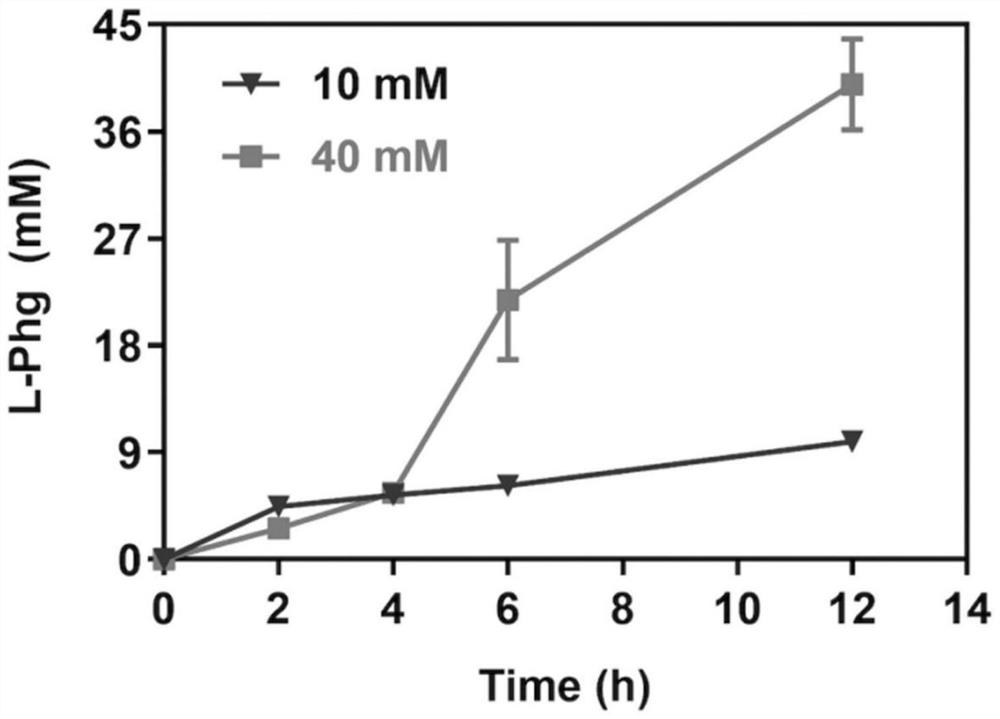
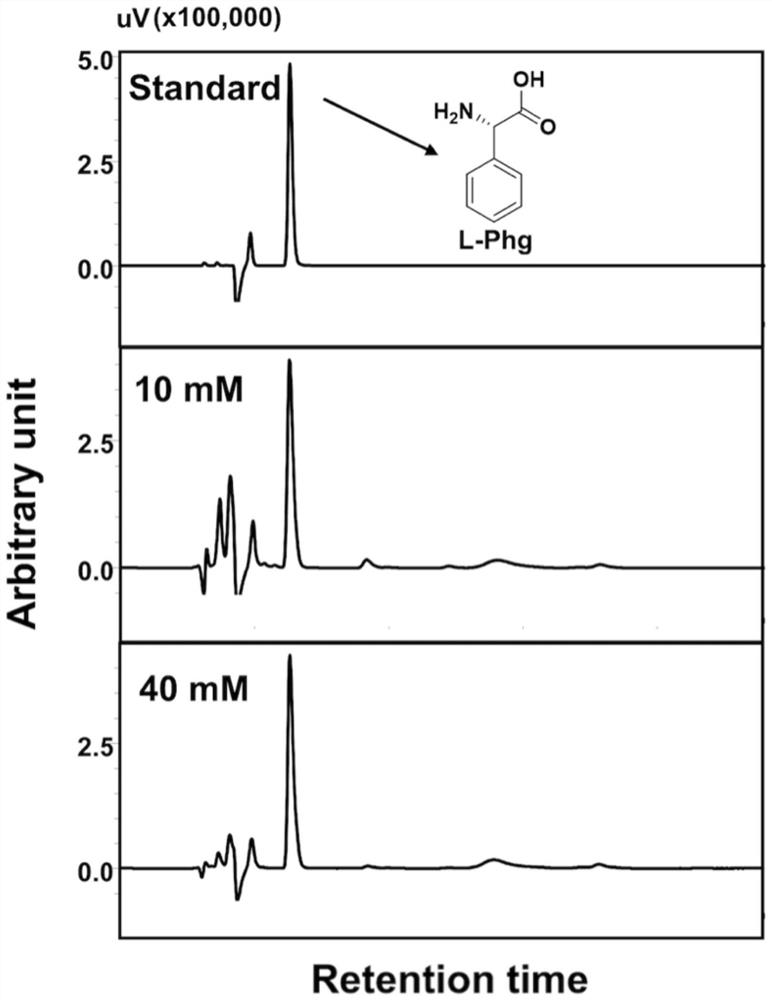
[0049] Whole cell transformation system (2mL): 10g / L dry weight of cells, L-phenylalanine (10mM or 40mM), and finally add phosphate buffer (200mM, pH=8.0) to make up. The system was reacted at 30° C. and 250 rpm for 1 to 12 hours. Then, the sample was centrifuged, the supernatant was taken, and the output of L-phenylglycine was detected by liquid chromatography.
[0050] Quantitative analysis of the product: the conversion solution was detected and analyzed by Shimadzu high performance liquid chromatography, using photodiode array detector (working wavelength 210nm), S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com