A kind of engineering bacteria for producing recombinant aspartic protease and its application
An amino acid, silkworm chrysalis protein technology, applied in the application, genetic engineering, recombinant DNA technology and other directions, can solve the problems of low expression, low efficiency, limited production of aspartic protease, etc., and achieves improved hydrolysis degree, improved solubility, excellent The effect of hydrolysis potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Cloning of aspartic protease encoding gene
[0031] Using Af293 derived from Aspergillus fumigatus Af293 as the parent, PCR was used to amplify the aspartic protease encoding gene SEQ ID NO.
Embodiment 2
[0032] Example 2 Preparation of aspartic protease
[0033] The expression vector pPIC9γ was double digested (EcoR I+Not I), and the gene encoding aspartic protease was double digested (EcoR I+Not I), and then the mature high specific activity xylan was digested The gene fragment of the enzyme mutant was connected with the expression vector pPIC9γ to obtain a recombinant plasmid containing the high specific activity xylanase mutant gene and transformed into Pichia pastoris GS115 to obtain a recombinant yeast strain.
[0034] Take the GS115 strain containing the recombinant plasmid, inoculate it into a 1L Erlenmeyer flask of 300mL BMGY medium, and place it at 30°C, 220rpm shaker for 48h; then centrifuge the culture solution at 3000×g for 5min, discard the supernatant, and use 100mL of 0.5 Resuspend in BMMY medium with % methanol, and place it again at 30°C, under the condition of 220rpm to induce culture. 0.5 mL of methanol was added every 12 h to keep the methanol concentratio...
Embodiment 3
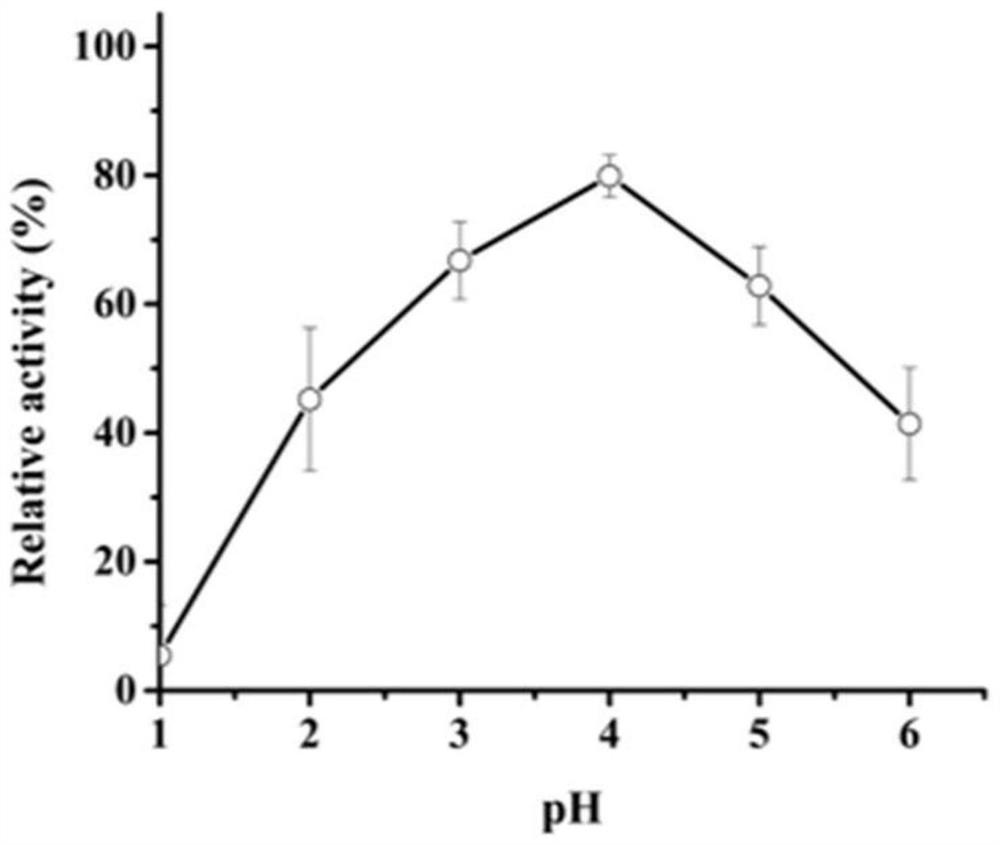
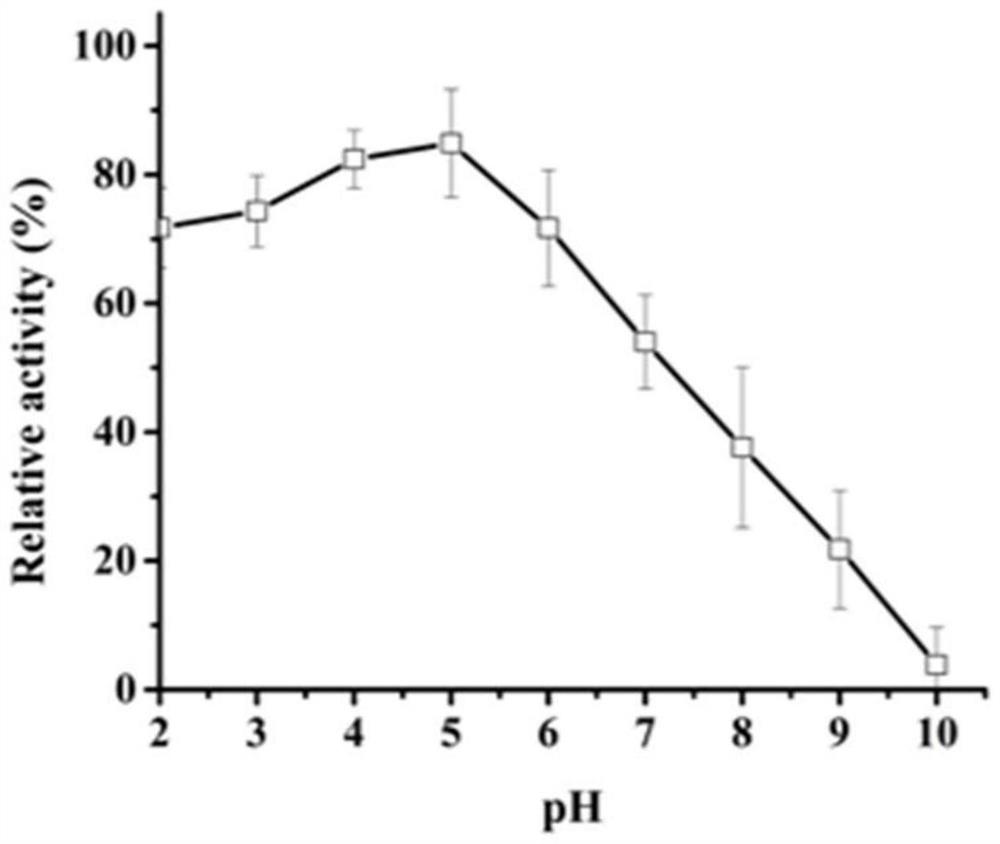
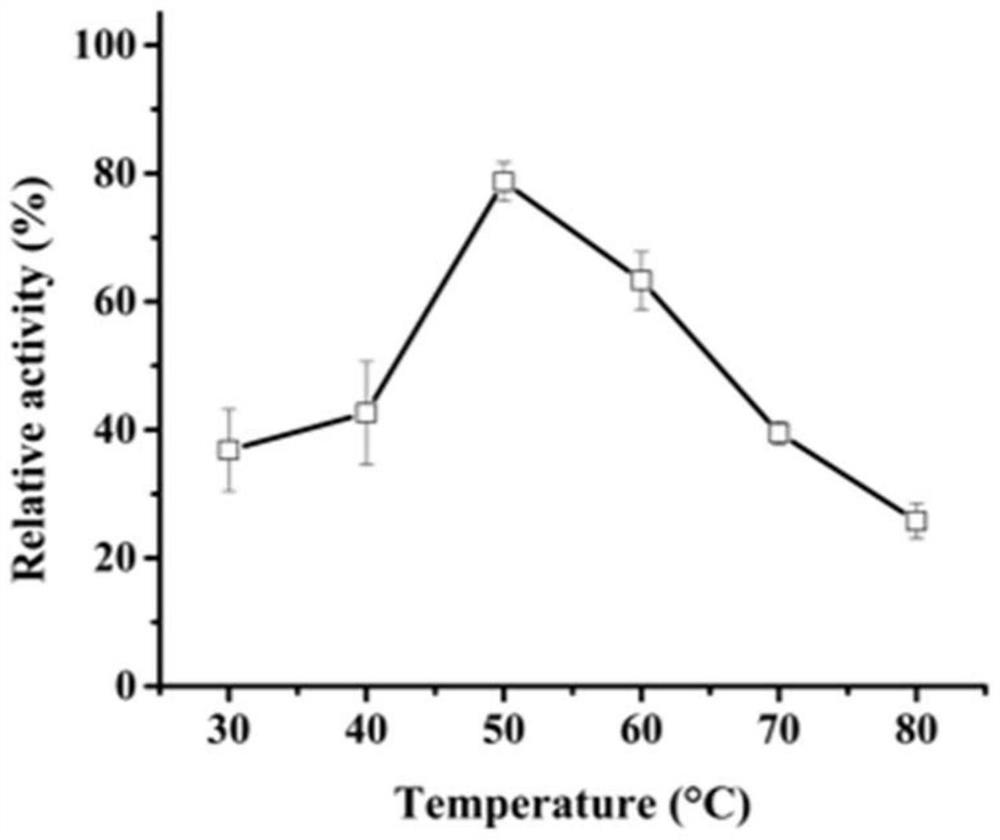
[0035] Example 3 Activity analysis of recombinant aspartic protease
[0036] Casein method: The specific method is as follows: The protease activity of AF293 was determined using casein as a substrate. Under the given pH and temperature conditions, 1 mL of reaction system consists of 500 μL of 1% casein and 500 μL of enzyme solution, and incubated for 10 minutes. The reaction was stopped by adding 1 mL of 10% trichloroacetic acid (TCA), centrifuged at 12,000 x g for 5 min, and then the supernatant (500 μL) was added to 2.5 mL of 0.4 M Na 2 CO 3 To the composed test tube, Folin and Ciocalteu reagent (500 μL) were added last. The mixture was further incubated at 40°C for 20 minutes and the absorbance was measured at 680nm.
PUM
| Property | Measurement | Unit |
|---|---|---|
| catalytic efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com