Primer combination and method for detecting genotype of high oleic acid peanut and application
A primer combination and primer set technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve the problems of cumbersome and complicated operations, high detection costs, and low accuracy, and achieve flexible throughput , good platform compatibility and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1, the design of the functional gene marker STARP primer of peanut high oleic character
[0086] The genes involved in high oleic peanut oleic acid content were AhFAD2A and AhFAD2B and their mutant alleles AhFAD2Asnp and AhFAD2Bindel.
[0087] 1. Acquisition of allelic variant sequences of high oleic acid peanut functional genes AhFAD2A and AhFAD2B
[0088] From the literature "Expressed variants of Δ12-fatty acid desaturase for the highleate trait in spanish market-type peanut lines. López Y, Nadaf HL, Smith OD, Simpson C, Fritz A. (2004) Molecular Breeding, 9:183-192." AhFAD2A and AhFAD2B and their mutant alleles AhFAD2Asnp and AhFAD2Bindel were obtained from .
[0089] 2. Determine the mutation sites of the allelic variation of the AhFAD2A and AhFAD2B genes
[0090] The nucleotide sequences of the coding regions of the AhFAD2A and AhFAD2Asnp genes were compared using the MEGA-X software. The 448th base of the AhFAD2A gene is G, while the 448th base of th...
Embodiment 2
[0100] Example 2, Establishment of STARP Marking Method for High-Oleic Peanut Genotype Detection
[0101] 1. Extraction of tested peanut material and its genomic DNA
[0102] (1) Selection of peanut materials for testing
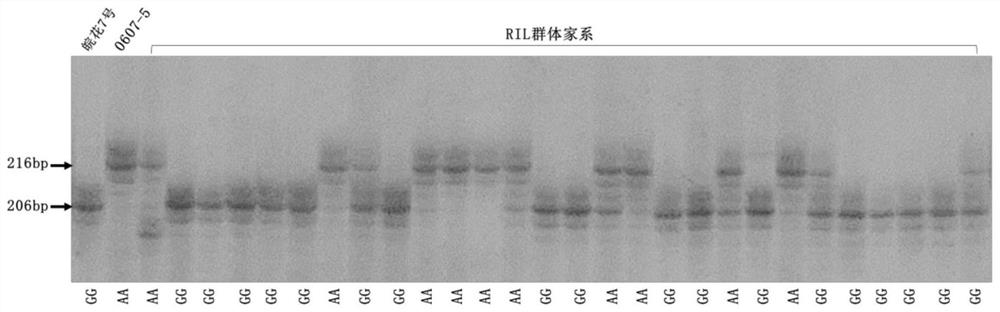
[0103] The peanut materials for testing in the present invention are high oleic acid peanut line 201731-1933 and common oleic acid content peanut line 201521-2612, wherein the AhFAD2A gene of 201731-1933 has G448A replacement, the genotype is AA homozygous type, and the AhFAD2B gene has 441_442InsA Mutation, the genotype is InsA homozygous type; AhFAD2A gene 448G and AhFAD2B gene 441_442 in 201521-2612 are not mutated, and the genotype is GG homozygous type and Del type respectively.
[0104] (2) Extraction of genomic DNA
[0105] A. Take a small amount of young peanut leaves (about 1g) and place them in a mortar, grind them into powder with liquid nitrogen, add 700 μL of 2% CTAB and 0.2β-mercaptoethanol extraction buffer that has been preheated in a 65°C ...
Embodiment 3
[0127] Example 3, Application of the method for identifying the FAD2 genotype by STARP markers in high oleic acid peanut breeding
[0128] Use the STARP marker developed by the present invention to detect the genotype of the RIL population family of the offspring of the hybrid combination of high oleic peanuts and ordinary peanuts, use the gene sequencing results to verify the typing results, test the fatty acid content of the RIL population family by gas chromatography, and analyze peanut oleic acid The relationship between content and genotype.
[0129] 1. Selection of peanut materials for testing
[0130] The peanut materials used in the test are 218 RIL population families of the offspring of the "0607-5×Wanhua No. 7" hybrid combination. The population has formed an F7 generation family through self-crossing and single-seed transmission. The internal genetics of the family are stable and the characters are uniform.
[0131] 2. Genomic DNA extraction
[0132] For extracti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com