Engineering bacterium containing nCas3 single-stranded endonuclease, preparation method and application
A single-stranded nucleic acid, engineering bacteria technology, applied in the biological field, can solve the problem of only reaching the editing efficiency, and achieve the effect of improving the transformation efficiency and editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Preparation and function of protein and its coding gene.
[0046] nCas3 single-stranded endonuclease refers to the introduction of mutations in the helicase functional domain of the wild-type Cas3 protein, making it lose the helicase activity, thereby transforming into nCas3 protein with only single-stranded nuclease activity. The endonuclease activity can act on a single strand of the target DNA duplex by endonucleating 1,4-phosphodiester bonds.
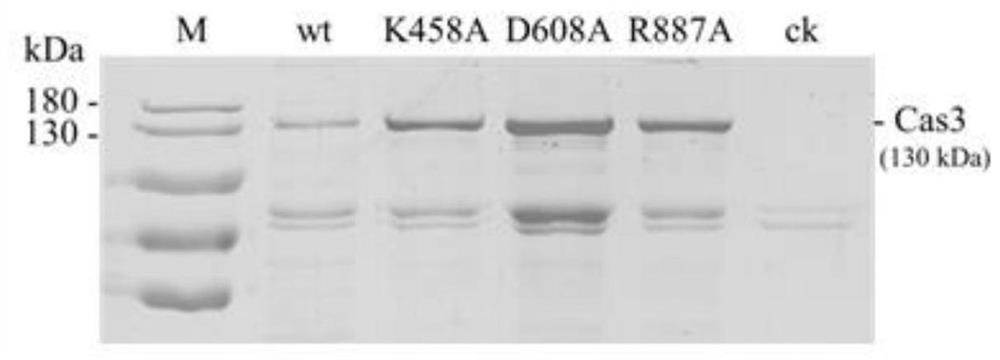
[0047] The amino acid sequence of the wild type (wide type) Cas3 protein involved in this embodiment of the present invention is shown in SEQ ID NO.1, and the nucleotide sequence is shown in SEQ ID NO.2. Three kinds of nCas3 single-stranded endonucleases with different sequences are specifically provided in this embodiment, named K458A respectively (the lysine at position 458 of the wild-type Cas3 protein is replaced with alanine, the amino acid sequence is the same as that of SEQ ID NO .1, the difference is only t...
Embodiment 2
[0070] Example 2 Using circular DNA as a substrate to verify protein function
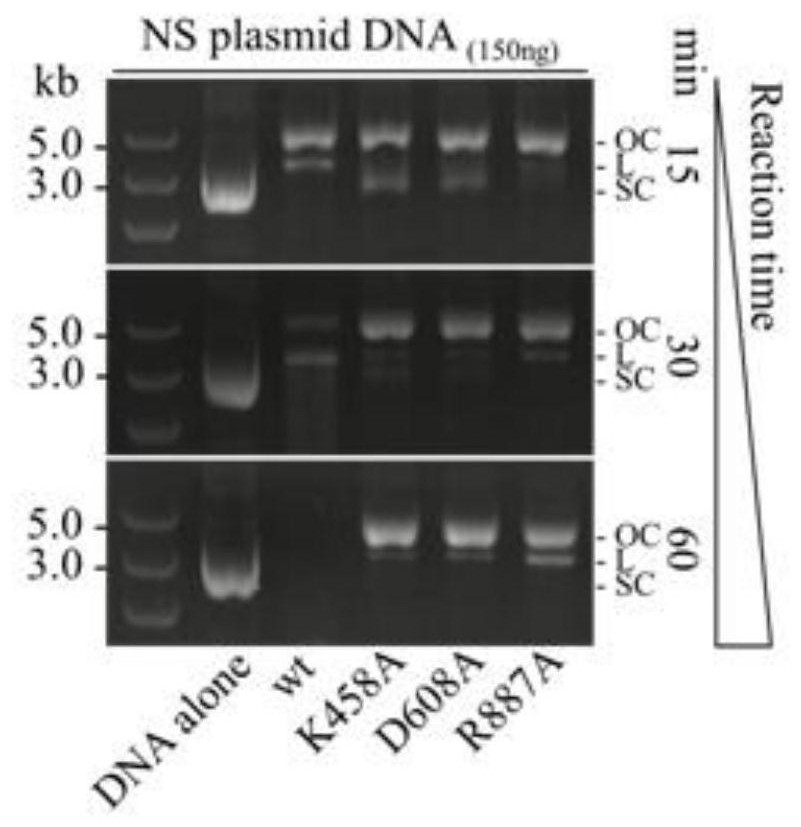
[0071] The pL2R plasmid (Zheng et al, 2019) was selected as the reaction substrate, and the total length of the plasmid was 3283bp. The reaction system contains 150ng pL2R circular plasmid; 2mM MgCl2; 0.5mM ATP; 250nM Cascade protein; 250nM one of the above purified Cas3 or nCas3 protein variants; 32nt crRNA. The above crRNA was synthesized by GenScript Biotechnology Co., Ltd., and its sequence is shown in SEQ ID NO.5. After reacting at 30°C for 15, 30, and 60 minutes respectively, the reaction products were run on agarose gel electrophoresis.
[0072] The result is as figure 2 as shown, figure 2 The leftmost lane in the center represents nucleic acid molecular weight standards (5.0, 3.0, 2.0kb); the rightmost Reaction time axis represents the reaction duration (15, 30, 60min); the right OC, L, SC represent circular DNA molecules after the reaction state, [OC (open circle); L (linear); SC (neg...
Embodiment 3
[0073] Embodiment 3 comprises the preparation of the engineering bacterium of nCas3 single-stranded endonuclease
[0074] In this example, D608A was taken as an example to prepare engineering bacteria containing nCas3 single-stranded endonuclease.
[0075] 1. Design and construct single base editing plasmid
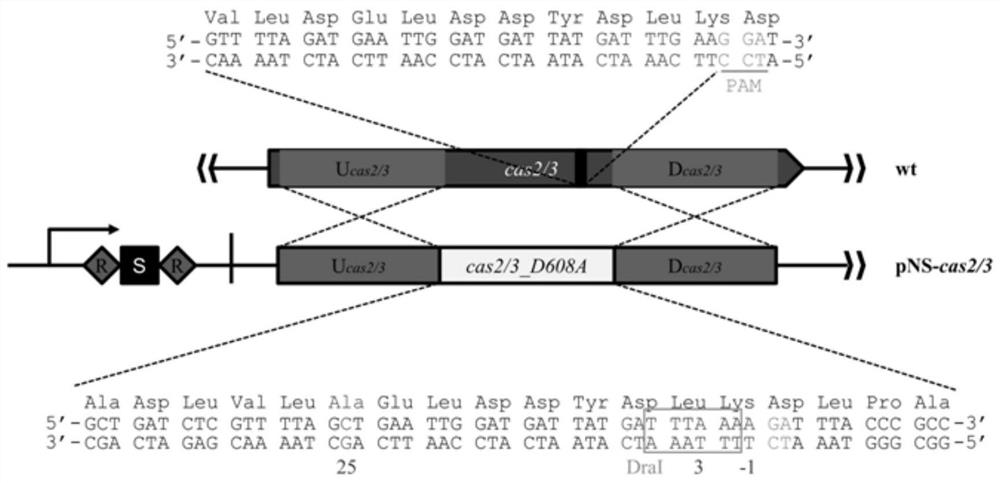
[0076] (1) According to the experimental requirements, it was decided to use the endogenous I-F type CRISPR-Cas editing system in the ZM4 strain to edit the target site on the genome.
[0077] Such as image 3 As shown, through careful analysis of the Cas3 protein coding gene on the genome of the ZM4 strain, it was found that there was a 5'-TCC-3' PAM sequence in the cas3 gene sequence, so the 32-nt sequence downstream of it was designed as a protospacer sequence . In order to prevent the CRISPR-Cas system from destroying the transferred donor plasmid, and to complete the in situ replacement of 676,661 adenine nucleotides to cytosine nucleotides in the ZMO0681 gene, a ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com