Tetracycline antibiotic degrading bacterium at low temperature and application thereof
A technology of tetracyclines and degrading bacteria, applied in the fields of applications, bacteria, microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Isolation of Pseudomonas putida
[0036] It is obtained from the soil of a livestock and poultry farm in Shenbei New District, Shenyang City, Liaoning Province through enrichment, separation and purification. Specifically, the following steps are included:
[0037] Step 1: Enrichment of strains
[0038] Weigh 5g of soil and add it to 250mL enrichment medium, which contains 20mg / L oxytetracycline, and incubate in the dark at 15°C and 160r / min in a shaker for 7 days. Take 5 mL of enriched culture medium and add it to a new enriched medium to continue culturing according to the above culture conditions, and then transfer again, and the concentration of antibiotics added to the enriched medium for each transfer is increased by 20 mg / L. Repeat the above steps for transfer culture until the concentration of antibiotics in the enriched medium reaches 100 mg / L.
[0039] Step 2: Isolation and purification of strains
[0040] Dilute the enriched culture solution wit...
Embodiment 2
[0047] Embodiment 2: bacterial strain 16S identification
[0048] (1) Genome extraction
[0049] Genomic DNA of strain TU-3 was extracted using the TIANamp Bacteria DNA Kit Bacterial Genomic DNA Extraction Kit from Tiangen Biochemical Technology Co., Ltd. and following the instructions.
[0050] (2) PCR amplification
[0051] The 16S universal primers are: 27F: AGAGTTTGATCCTGGCTCAG 1492R: TACGGCTACCTTGTTACGACTT, and the PCR length is about 1500bp.
[0052] PCR reaction system:
[0053]
[0054] PCR reaction program:
[0055] Pre-denaturation at 95°C for 4min, deformation at 94°C for 30s, annealing at 55°C for 30s, extension at 72°C for 40s, 35 cycles, extension at 72°C for 7min, and 4°C until termination.
[0056] (3) Gel electrophoresis
[0057] Finally, the obtained PCR product was observed by 1% agarose gel electrophoresis, and the electrophoresis condition was 80V for 30min. The PCR products that were successfully amplified were sent to Huada Gene Technology Co., ...
Embodiment 3
[0063] Example 3: Verification of the degradation effect of TU-3 on oxytetracycline
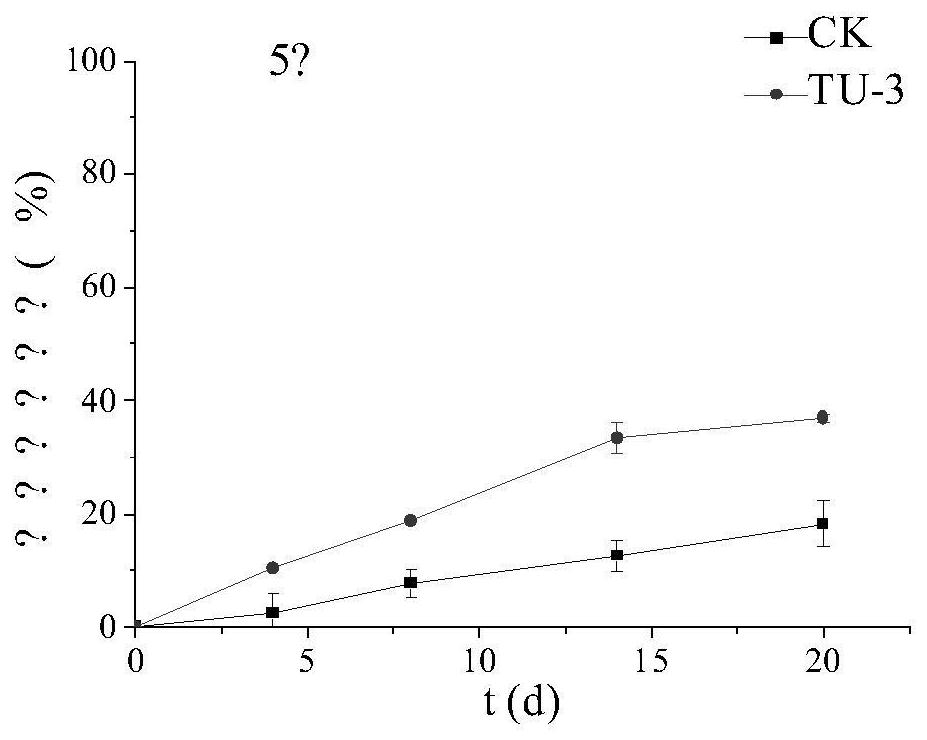
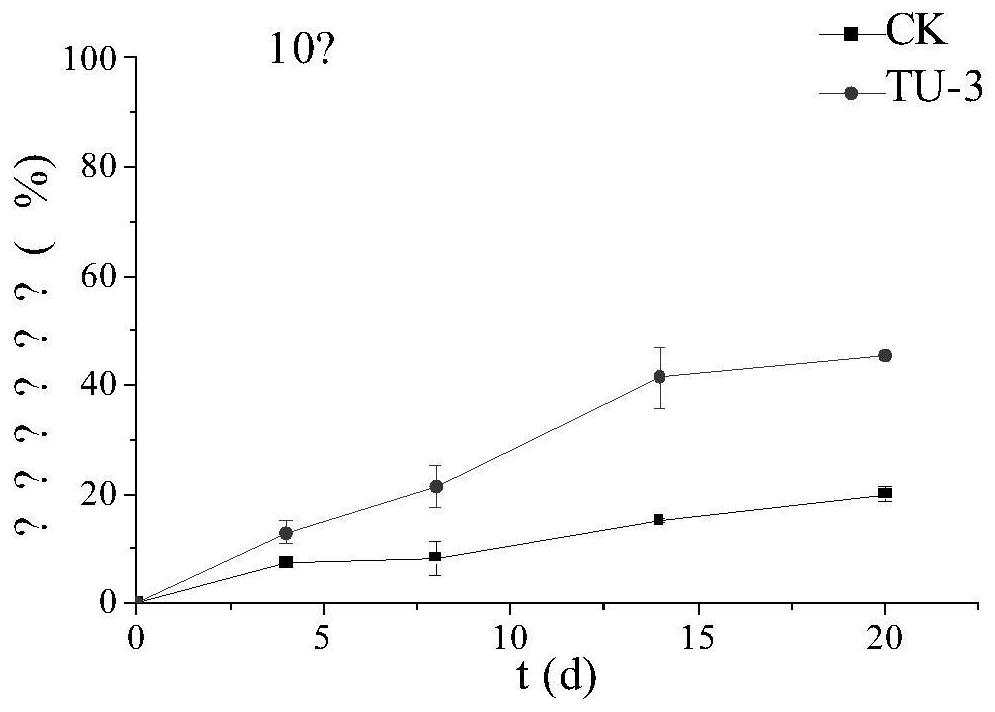
[0064] Shake culture TU-3 in LB medium at 37°C and 160rpm for 24 hours, then inoculate 5% of the inoculum into the selection medium containing oxytetracycline 20 mg per liter, and culture at 5°C and 10°C respectively. And 15 ℃, 160rpm conditions of shaking culture in the dark for 20d, each group set up three parallel, with no bacteria as a blank control. The resulting culture was passed through a 0.22um organic filter, and the residual concentration of oxytetracycline was detected on the machine.
[0065] Calculated as follows:
[0066]
[0067] The results showed that the initial content of oxytetracycline was 20mg / L, and the degradation rate of the bacteria could reach 66.6% in 8 days at 15°C and 160rpm, and the degradation rate of the blank control was 32%; at 10°C and 160rpm, the degradation rate in 20d could reach 45.5%. %, the degradation rate of the blank control is 20%; under the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com