Multivalent aptamer functionalized DNA nano-structure probe as well as preparation method and application thereof
A nanostructure and functionalization technology, applied in the field of biomedicine, achieves the effects of stable structure, improved binding efficiency, and simple preparation method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Multivalent aptamer functionalized DNA nanostructure probe and its preparation and structure characterization experiment
[0043] 1. Multivalent aptamer-functionalized DNA nanostructure probes
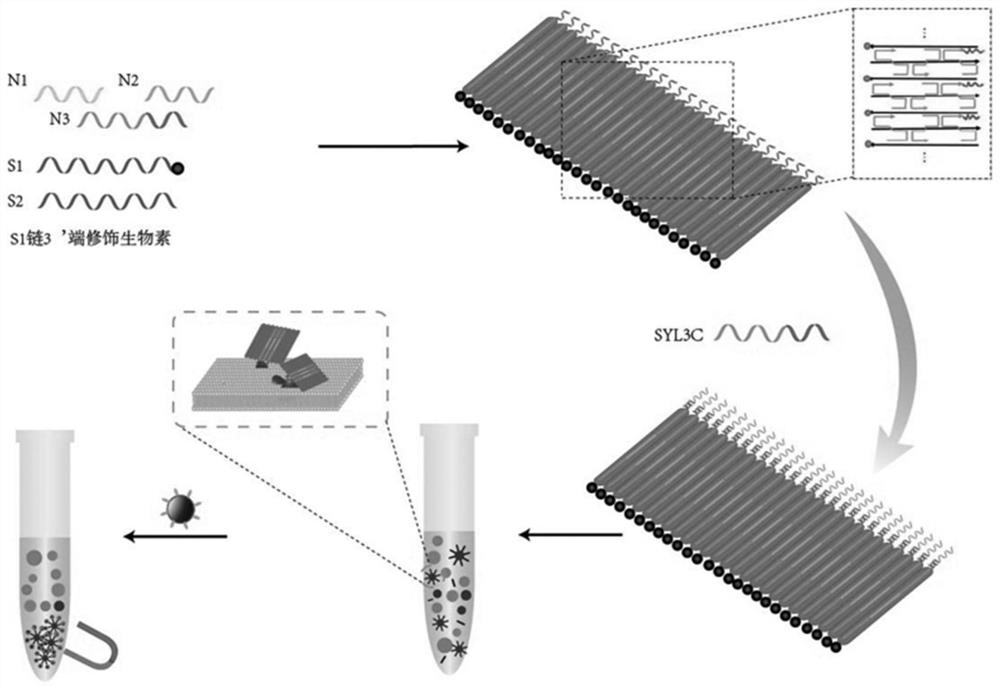
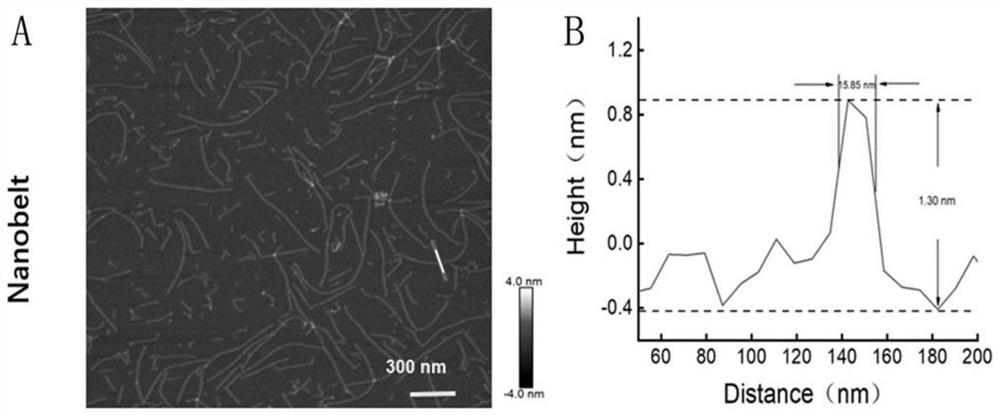
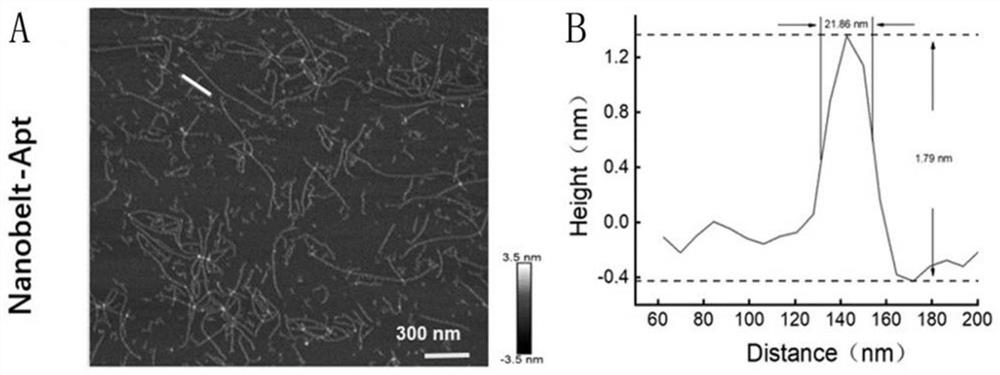
[0044] like figure 1As shown in the figure, one side of the DNA nanostructure of the probe has multiple DNA nucleic acid aptamers, which can specifically bind to protein receptors on the membrane surface of circulating tumor cells; the other side has multiple biotin sites, which can bind to streptavidin Avidin-modified magnetic particles; the probe is synthesized from a total of six DNA single strands of S1, S2, N1, N2, N3, and SYL3C, as follows:
[0045] Described S1 is shown as SEQ ID NO.1, specifically:
[0046] 5'-CAGGGCTGGGCATAGAAGTCAGGGCAGAGACGAGTTGAGAATACGAGT-TEG-Biotin-3';
[0047] The S1 is a sequence of modified biotin molecules, which can be combined with streptavidin-modified magnetic particles.
[0048] Described S2 is as shown in SEQ ID NO.2, specific...
Embodiment 2
[0066] Using the multivalent aptamer-functionalized DNA nanostructure probe prepared in Example 1 to realize the capture of human breast cancer cells (MCF-7) and the cell number gradient experiment, the details are as follows:
[0067] (1) MCF-7 cell staining: The passaged MCF-7 cells were placed at 37°C in 5% CO containing 2 After 48h, use 1×PBS (containing NaCl, KCl, Na 2 HPO 4 , KH 2 PO 4 , pH=7.4) to wash, remove the washing liquid, add DMEM without fetal bovine serum (FBS), and 5 μL of cell membrane dye DIO (1 mM), mix well and place in a cell incubator for staining for 20 min.
[0068] (2) MCF-7 cell digestion: After staining, remove the stain from the culture flask and wash with 1×PBS at least three times to avoid excess stain affecting cell viability, add trypsin (containing ethylenediaminetetraacetic acid (EDTA) EDTA) 0.02%), place the culture flask in the cell culture incubator again for about 2 min, then supplement with FBS-containing high-glucose medium DMEM, s...
Embodiment 3
[0080] The multivalent aptamer functionalized DNA nanostructure probe prepared in Example 1 was used to realize the capture of human pancreatic cancer cells (BxPC-3) and the cell number gradient experiment, as follows:
[0081] Referring to the operation of capturing MCF-7 with the multivalent aptamer-functionalized DNA nanostructure probe in Example 2, the difference in this example is that the cultured cells are BxPC-3, and the rest of the steps are the same as those in Example 2.
[0082] Experimental results: In this example, the capture efficiency of the multivalent aptamer-functionalized DNA nanostructure probe to capture BxPC-3 is as follows: Figure 5 As shown, the average capture efficiency of BxPC-3 was 52.9%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com