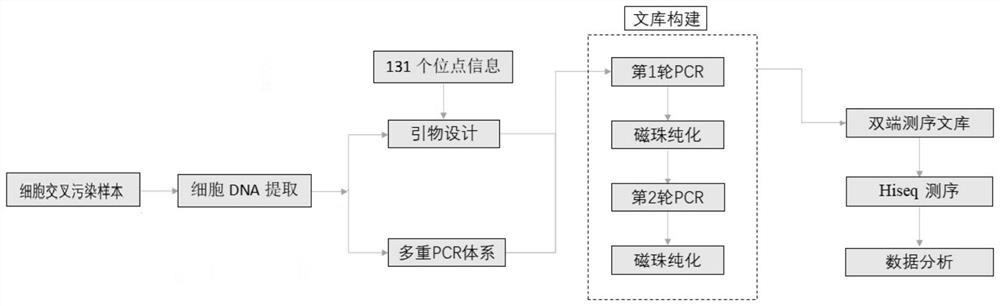
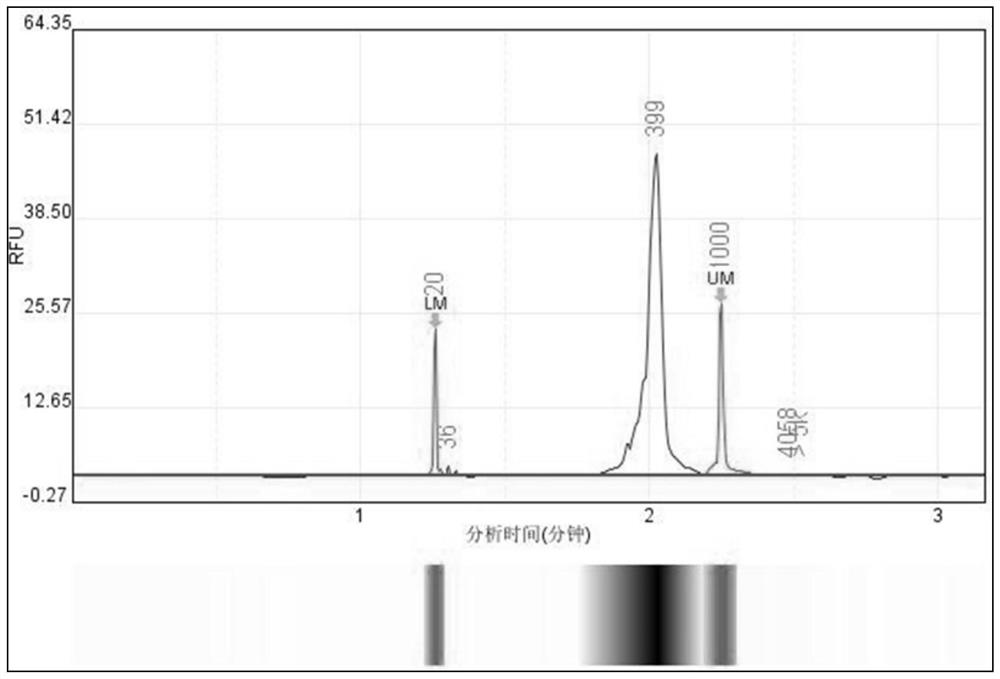
Cell cross contamination detection method and application thereof
A technology of cross-contamination and detection method, applied in the field of cell cross-contamination detection, can solve the problems of contamination and insufficient analysis ability of complex mixed cell system, and achieve the effects of high sensitivity, low allele coverage and high specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Simulated detection of cell cross-contamination based on mixed cell samples
[0043] 1. Cell culture
[0044] The DMEM medium containing 10% fetal bovine serum was used, and the cells were cultured in an incubator containing 5% carbon dioxide at 37° C., and the cells could be used after they had grown into monolayers.
[0045] 2. Simulate cell cross-contamination:
[0046] After the adherent cells are digested with trypsin, a single cell suspension is prepared, the corresponding cell concentration is determined, and the required corresponding single cell suspension volume is calculated according to the specific number of cells. (1:9, 1:19, 1:49, 1:99) were mixed and used as test samples for simulated contamination of cells.
[0047] 3. DNA extraction from simulated contaminated cells:
[0048] DNA extraction kits were used to extract the DNA of simulated contaminated cells according to the instructions, and the concentration was measured with a micro-nuclei...
Embodiment 2
[0064] Example 2 Simulated detection of cell cross-contamination based on mixed cell DNA samples
[0065] 1. Cell culture:
[0066] The DMEM medium containing 10% fetal bovine serum was used, and the cells were cultured in an incubator containing 5% carbon dioxide at 37° C., and the cells could be used after they had grown into monolayers.
[0067] 2. Cell DNA extraction:
[0068] The adherent cells were digested with trypsin to prepare a single-cell suspension; then, according to the instructions of the DNA extraction kit, the DNA of each cell was extracted separately, and the concentration was determined with a micro-nucleic acid protein detector.
[0069] 3. Simulate cell cross-contamination:
[0070] Determine the dilution ratio according to the measured DNA concentration, and dilute the extracted DNA from each cell to a concentration of 10ng / μL. Then, the extracted DNAs from different cells were mixed in different ratios (1:9, 1:19, 1:49, 1:99) to simulate cell cross-c...
Embodiment 3
[0079] Example 3 Comparison between the method of the present invention and the STR typing method
[0080] 1. Cell culture:
[0081] The DMEM medium containing 10% fetal bovine serum was used, and the cells were cultured in an incubator containing 5% carbon dioxide at 37° C. and used after the cells grew into a monolayer.
[0082] 2. Cell DNA sample preparation:
[0083] The cell line DNA extracted in the same batch in Example 1 was used, and mixed according to the ratio of different cell numbers (1:9, 1:19, 1:49, 1:99), as a detection sample for simulated cross-contamination of cells, and the sample concentration was ≥ 50ng / μL.
[0084] 3. STR analysis and identification:
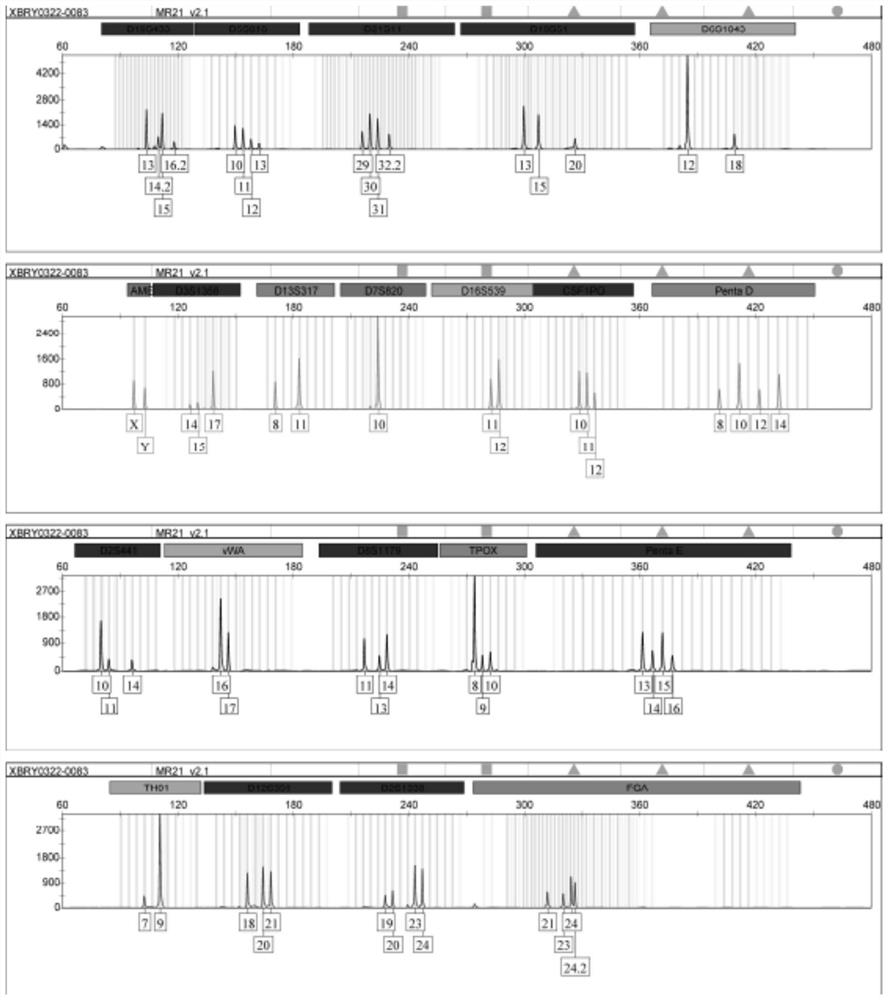
[0085] Take an appropriate amount of simulated cross-contaminated cell DNA, use MicroreaderTM21 ID System to amplify 20 STR loci and sex identification loci, and use ABI 3730xl genetic analyzer to detect PCR products.
[0086] 4. STR information comparison:
[0087] The detection data information was pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com