Method for producing L-ornithine by using mixed sugar raw material in double-bacterium co-culture system
A co-cultivation system, ornithine technology, applied in the field of genetic engineering and biology, to achieve the effect of expanding the substrate spectrum, reducing production costs and optimizing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Xylose Utilizing Strain Corynebacterium glutamicum Construction of GX01
[0040] (1) Construction of integrated plasmid: Escherichia coli Escherichia coli The MG1655 genome was used as a template and upstream primers were used xyl AB-F: atcgaaaatgcaagcctattttgaccagct (SEQ. No. 1) and downstream primers xyl AB-R: ccaaagatttacgccattaatggcagaagttgc (SEQ.No.2) PCR amplification xyl AB fragment (such as SEQ.No.3 in the Sequence Listing xyl AB sequence shown). The PCR conditions were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 62°C for 30 s, extension at 72°C for 3 min, final extension at 72°C for 5 min, and storage at 4°C for 59 min; the number of cycles was 30 cycles.
[0041] Corynebacterium glutamicum C. glutamicum jp-07092 genome as template, using primers ldh A-up-F1: cggtacccggggatcggaacaccatgcgattaaggtgc (SEQ. No. 4) and ldh A-up-R1: PCR amplification of gcttgcattttcgatcccacttcctgattt cc (SEQ.No.5) ldh...
Embodiment 2
[0050] Example 2: Arabinose Utilizing Strain Corynebacterium glutamicum Construction of GA01
[0051](1) Construction of integrated plasmid: Escherichia coli Escherichia coli The MG1655 genome was used as a template and upstream primers were used ara BAD-F: atcgaaaatggcgattgcaattggcc (SEQ. No. 10) and downstream primers ara BAD-R: ccaaagatttactgcccgtaatatgccttcgc (SEQ.No.11) amplified by PCR ara BAD fragment (such as SEQ.No.12 in the Sequence Listing ara BAD sequence shown). The PCR conditions were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 5 min, final extension at 72°C for 5 min, and storage at 4°C for 59 min; the number of cycles was 30 cycles.
[0052] Corynebacterium glutamicum Corynebacterium glutamicum The jp-07092 genome was used as a template, using ldh A-up-F2: cggtacccgg ggatcggaac accatgcgat taaggtgc (SEQ. No. 13) and ldh A-up-R2: atcgccattt tcgatcccac ttcctgatt (SEQ.No.14) a...
Embodiment 3
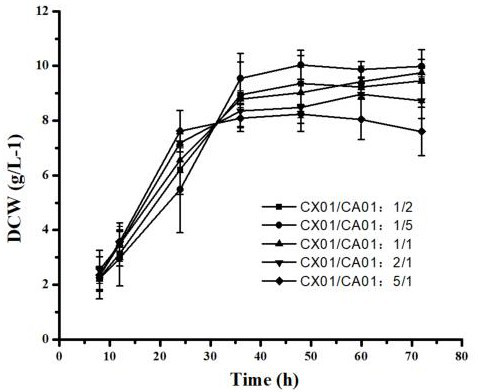
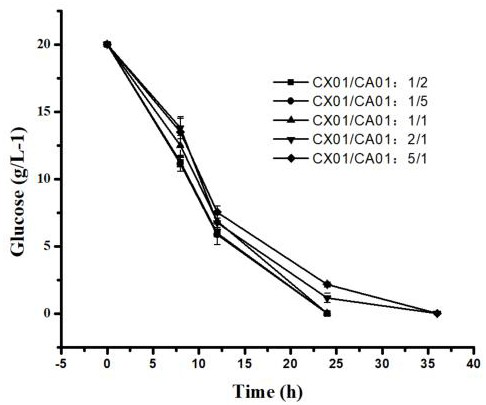
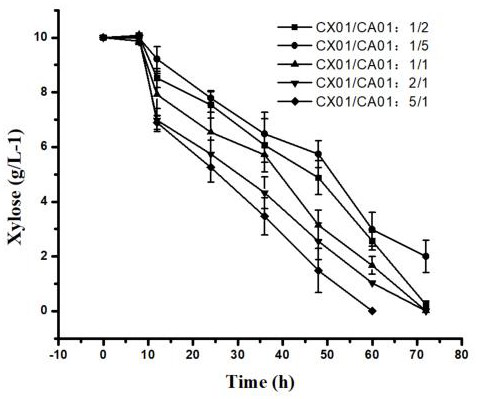
[0061] Example 3: A Utilization Corynebacterium glutamicum GX01 and Corynebacterium glutamicum Method for producing L-ornithine by using mixed sugar raw material in GA01 double bacteria co-cultivation system
[0062] (1) Construction of Corynebacterium glutamicum Corynebacterium glutamicum GX01 and Corynebacterium glutamicum Dual bacteria co-culture system of GA01
[0063] Prepare LB solid medium: tryptone 10 g / L, yeast powder 5 g / L, sodium chloride 10 g / L, agar powder 20 g / L, sterilize at 121 °C for 20 min.
[0064] Preparation of seed liquid medium: glucose 20 g / L, K 2 HPO 4 ·3H 2 O 1.5 g / L, KH 2 PO 4 0.5 g / L, MgSO 4 ·7H 2 O 0.4 g / L, urea 2.5 g / L, MnSO 4 ·H 2 O 0.02 g / L, FeSO 4 ·7H 2 O 0.02 g / L, Biotin 0.1 mg / L, VB 1 0.1 mg / L, arginine 0.02 g / L, with H 3 PO 4 Adjust pH to 7.0 and sterilize at 110 °C for 10 min.
[0065] The formula for preparing the co-culture fermentation medium is: glucose 20 g / L, arabinose 20 g / L, xylose 10 g / L, K 2 HPO 4 ·3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com