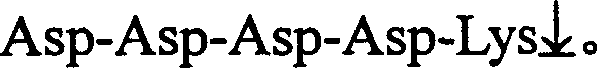Enterpeptidase light chain variant with high activity and high stability
A high-stability, enterokinase technology, applied in the field of serine proteolytic enzymes, can solve the problems of decreased enzyme activity, decreased enzyme activity, pollution, etc., and achieves the effects of low cost and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0016] 1. Preparation of duodenal total RNA, fresh bovine or human duodenal tissue was taken, and total RNA was extracted according to the operation manual of Qigen kit (Catalog NO.74104).
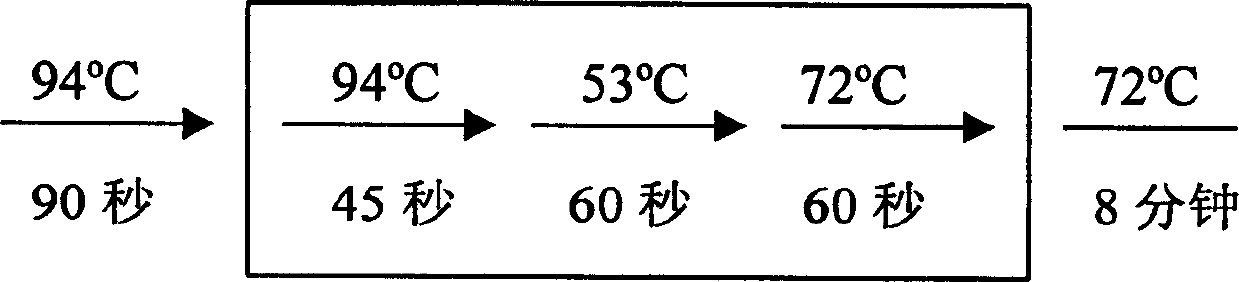
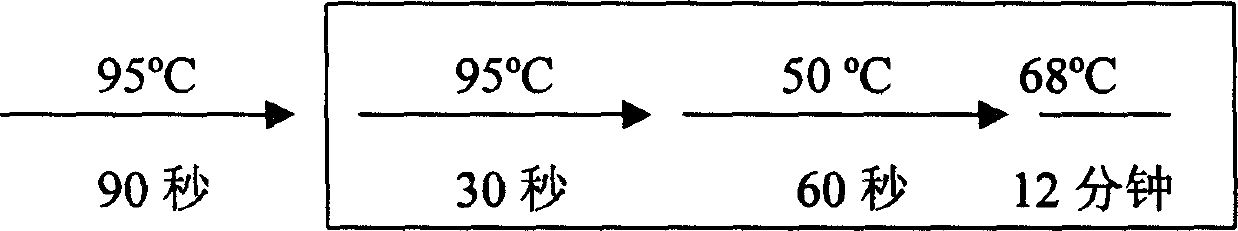
[0017] 2. RT-PCR reaction
[0018] The RT-PCR kit was purchased from Invitrogen Corporation (Catalog No. L1310-01). The experimental process refers to the operation manual. Take 3 μg of total RNA, add water to a final volume of 11.5 μl, then add 1 μl thymine oligonucleotide primer, mix well, incubate at 65°C for 10 minutes, room temperature for 2 minutes, then add 1 μl RNase inhibitor, 4 μl 5x reaction buffer, 1 μl 100mM dNTP, 1μl 80mM sodium pyrophosphate, 0.5μl AMV reverse transcriptase, mix well and set it at 42°C for 60min.
[0019] Take 3 μl of the above reaction solution as a template for PCR amplification. When prokaryotic cells are used as expression host bacteria, the 5' end primers for PCR amplification are CAT ATG GAC GAC GAT GAC AAG ATT GTC GGA GGA AGT GAC TCC , the 3′ end...
Embodiment 1
[0058] Fermentation with BL21 engineered bacteria of enterokinase light chain variant. Taking a 10-liter fermenter as an example (fermentation lasts for 2 days), 27.8 grams of bacteria can be obtained per liter of fermentation broth, containing about 578 mg of enterokinase light chain variant protein, and 415 mg of denatured enterokinase protein is obtained by Zn-Sepharose affinity chromatography. Kinase light chain variant protein, enterokinase activity 2.19×10 per liter after renaturation treatment 5 u, after STI-Sepharose affinity chromatography, the recovery of enterokinase activity was 1.64×10 5 u, get 1.64×10 per 10 liters of fermentation broth 6 uEnterokinase activity. Calculated by hydrolyzing 50ug of the fusion protein per unit of enterokinase activity, the enterokinase obtained per 10 liters of fermentation broth can degrade 82 grams of the fusion protein.
[0059] Using the above method to prepare wild-type enterokinase light chain protein, only 1.18×10 per 10 li...
Embodiment 2
[0061] Fermentation with yeast engineered strains of enterokinase light chain variants (fermentation time lasted 96 hours). Taking a 10-liter fermenter as an example, the enterokinase activity contained in the supernatant of each liter of fermentation broth is 1.45×10 6 u, after Zn-Sepharose affinity chromatography, the recovery of enterokinase activity was 1.03×10 6 u, 10 liters of fermentation broth obtained 1.03×10 7 uEnterokinase activity, calculated by hydrolyzing 50 μg of fusion protein per unit of enterokinase activity, the enterokinase obtained from 10 liters of fermentation broth can degrade 515 grams of fusion protein.
[0062] References: Grant, D.A.W. & Hermon-Taylar, J. Hydrolysis of artificial substrates by enterokinase and trypsin and the development of a sensitive specific assay for enterokinase in serum. Biochim. Biophys. Acta, (1979); 567, 207-215.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com