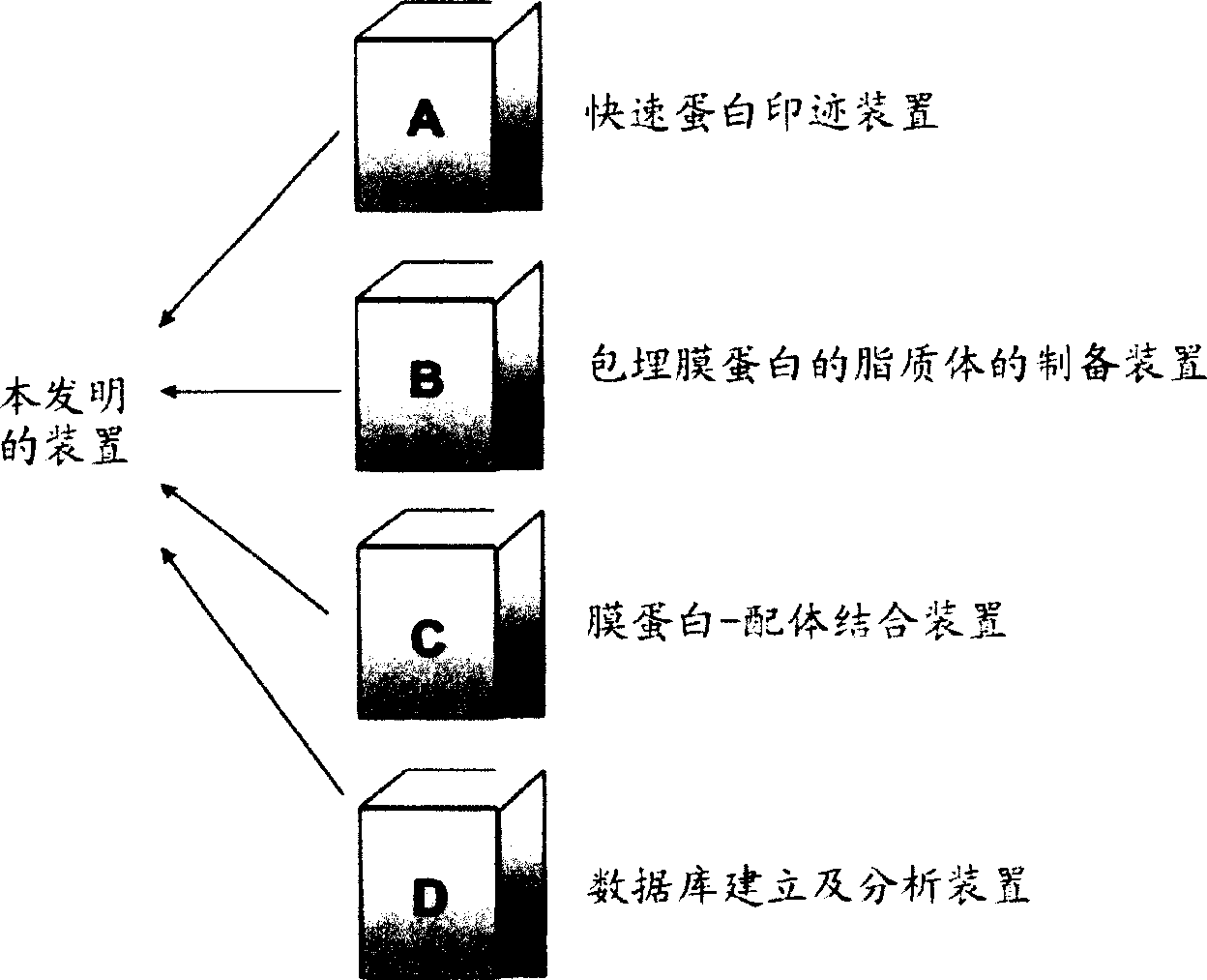
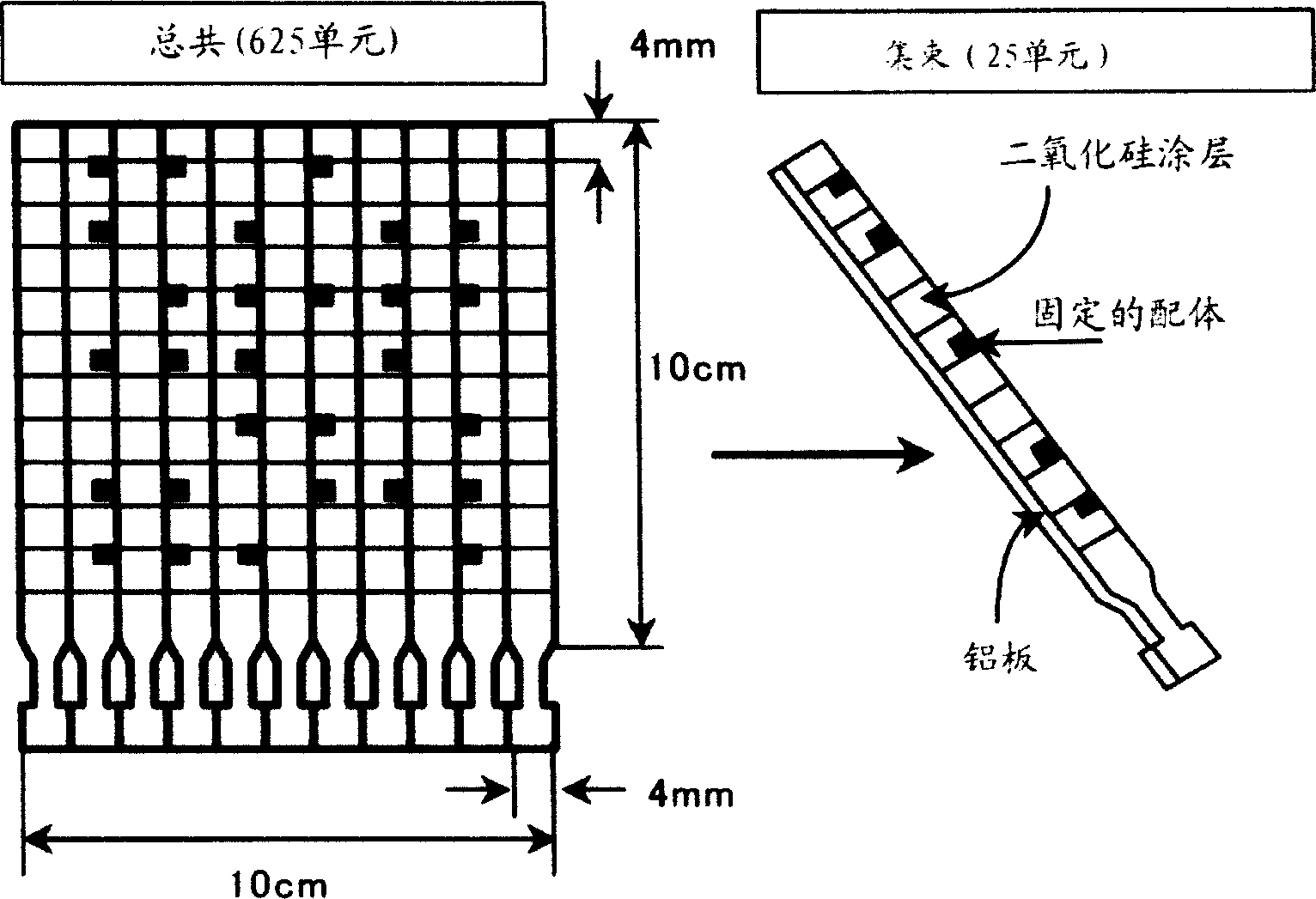
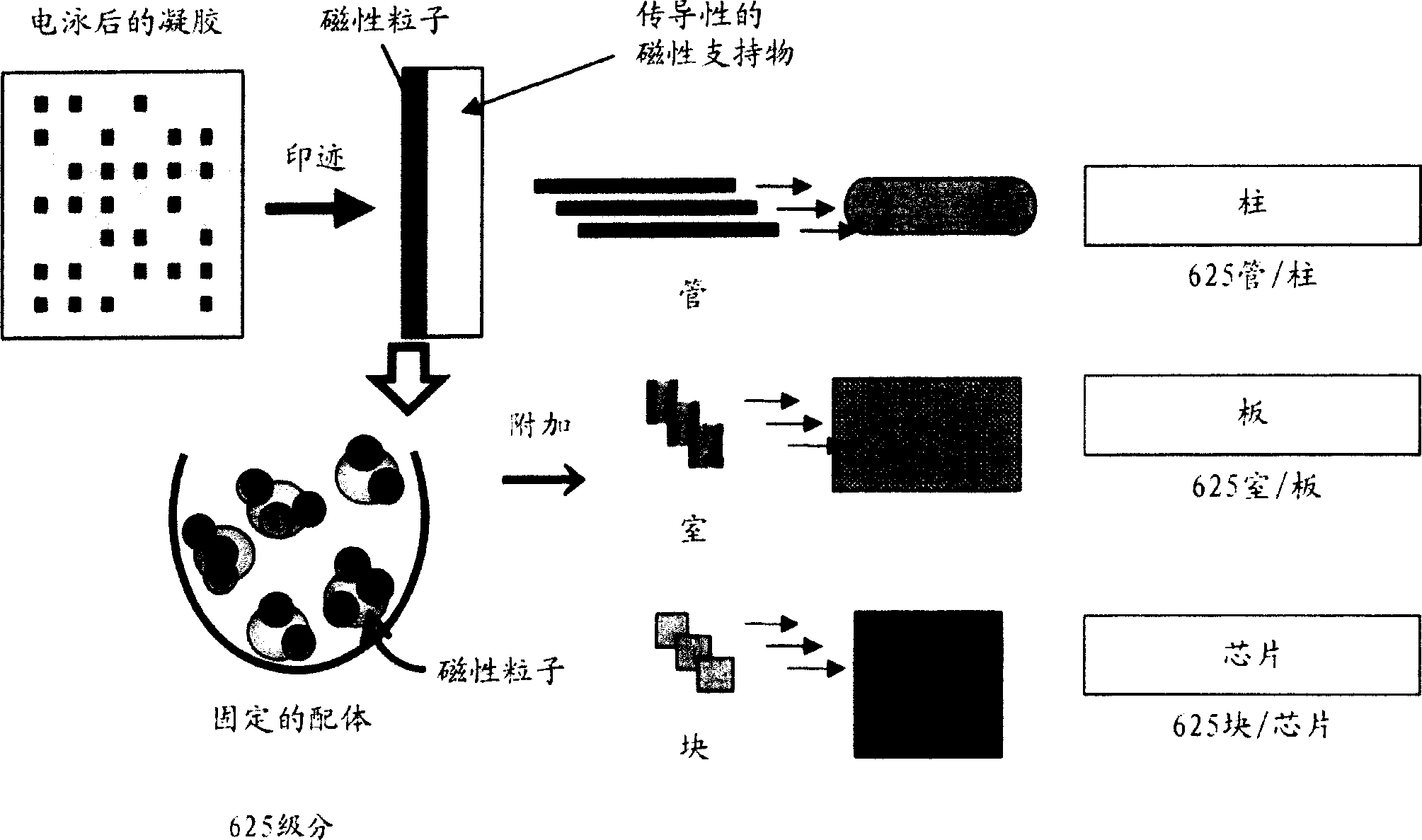
Novel proteome analysis method and devices therefor
A technology of proteomics and analysis methods, applied in the field of database itself, can solve the problems of backward purification and separation research, time-consuming, complex multi-step, shortening of detection time, etc., and achieve the effect of shortening operation time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0208] Preparation of Liposomes Encapsulating Urokinase Receptor
[0209] (1) Preparation of membrane fraction
[0210] Since U937 is a cell line derived from human monocytes and expresses a high concentration of urokinase receptor after being stimulated by phorbol ester (PMA), it was used as a sample of the isolated membrane fraction. After washing, cells were disrupted by treatment with Polytron 3 times at 1 min intervals for 2-5 sec each under ice-cooled conditions, while the membrane fraction was centrifuged through a 40% sucrose density gradient (95,000 g x 60 min) enriched on the interface ( Figure 6 ).
[0211] (2) Preparation of liposomes embedding membrane proteins
[0212] The purified yolk lecithin (1.25 g) and cholesterol (0.125 g) were suspended in 25 mL of physiological saline, and treated with a probe-type ultrasonic device for 15 minutes under ice-cooling. The average particle size of the obtained liposomes was 80 nm. Add the pre-prepared U937 membrane ...
Embodiment 2
[0223] Example 2: Appearance of receptor-embedded liposomes after particle diameter reduction
[0224] The particle size of liposomes embedded in membrane proteosomes was changed by extruder method, and the appearance of liposomes embedded in urokinase receptor was examined by fluorescent (FITC)-labeled urokinase. As a result, the number of liposomes embedding target receptors in the liposome solution increased significantly when the pore size of the filter passed through was not more than 0.6 μm, as shown in FIG. 16 . It is speculated to be caused by the fact that immediately after fusion the vast majority of target receptors are encapsulated in large liposomes in multilamellar liposomes, fluorescence cannot be detected, and more importantly, the smaller size of liposomes Plastids lead to a reduction in the number of receptors entrapped in a liposome, thus causing a marked difference between liposomes entrapping the target receptor and liposomes not entrapping the target rece...
Embodiment 3
[0226] Example 3: Blotting of Urokinase onto Mass Spectrometry Plates
[0227] Urokinase (10, 13, 16, 20, 33 μg) was loaded into the same well of the polyacrylamide gel at constant time intervals and electrophoresed. After migration was complete, the gel was peeled off from the glass plate, and after 5 minutes of replacing the solution in the gel with blotting buffer (5% acetonitrile / 125 mM NaCl / PBS), it was hot-blotted (diffused) onto an aluminum plate whose surface Treated with a hydrophobic substance having 16 carbon atoms. As the blotting buffer, phosphate buffer containing 5% acetonitrile and 125 mM NaCl was used for blotting at 35° C. for 4 hours. Figure 17 Shown are photographs of the mass spectrum plate after blotting and of gels stained with Coomassie Brilliant Blue before and after blotting. The urokinase band on the mass spectrometer plate cannot be seen with the naked eye, but since the urokinase band on the gel becomes weaker after blotting, it implies that a c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| The average particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com