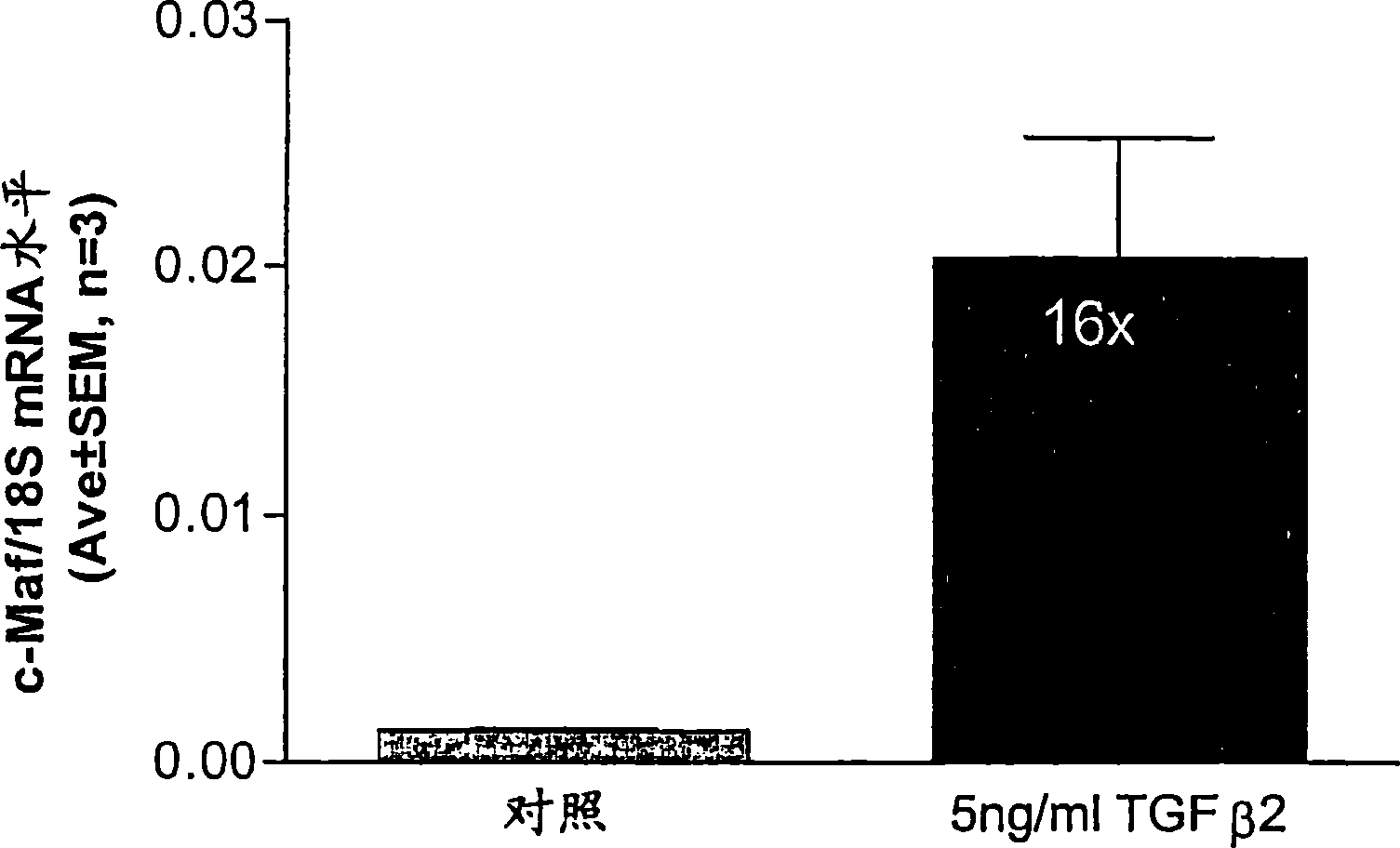
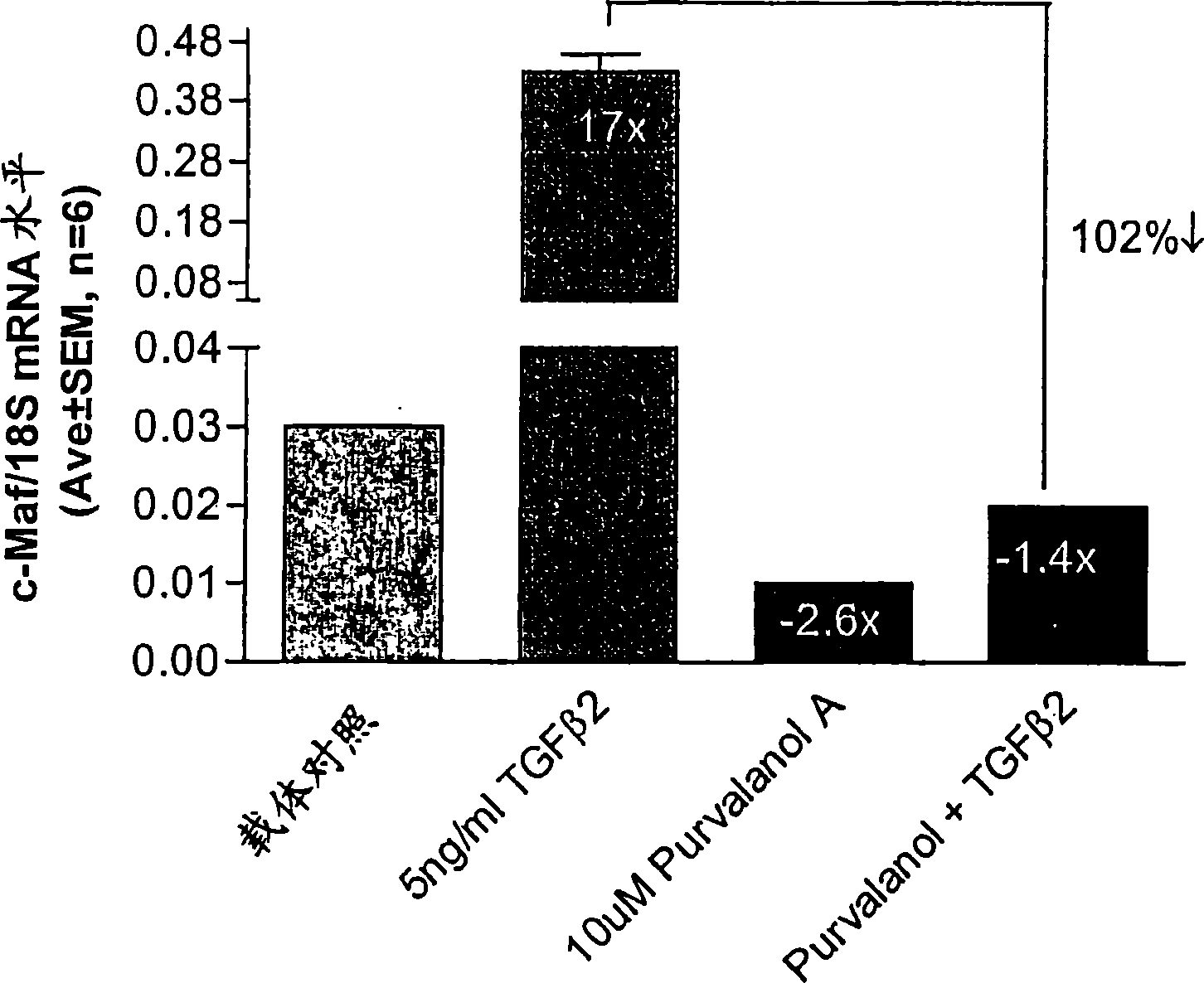
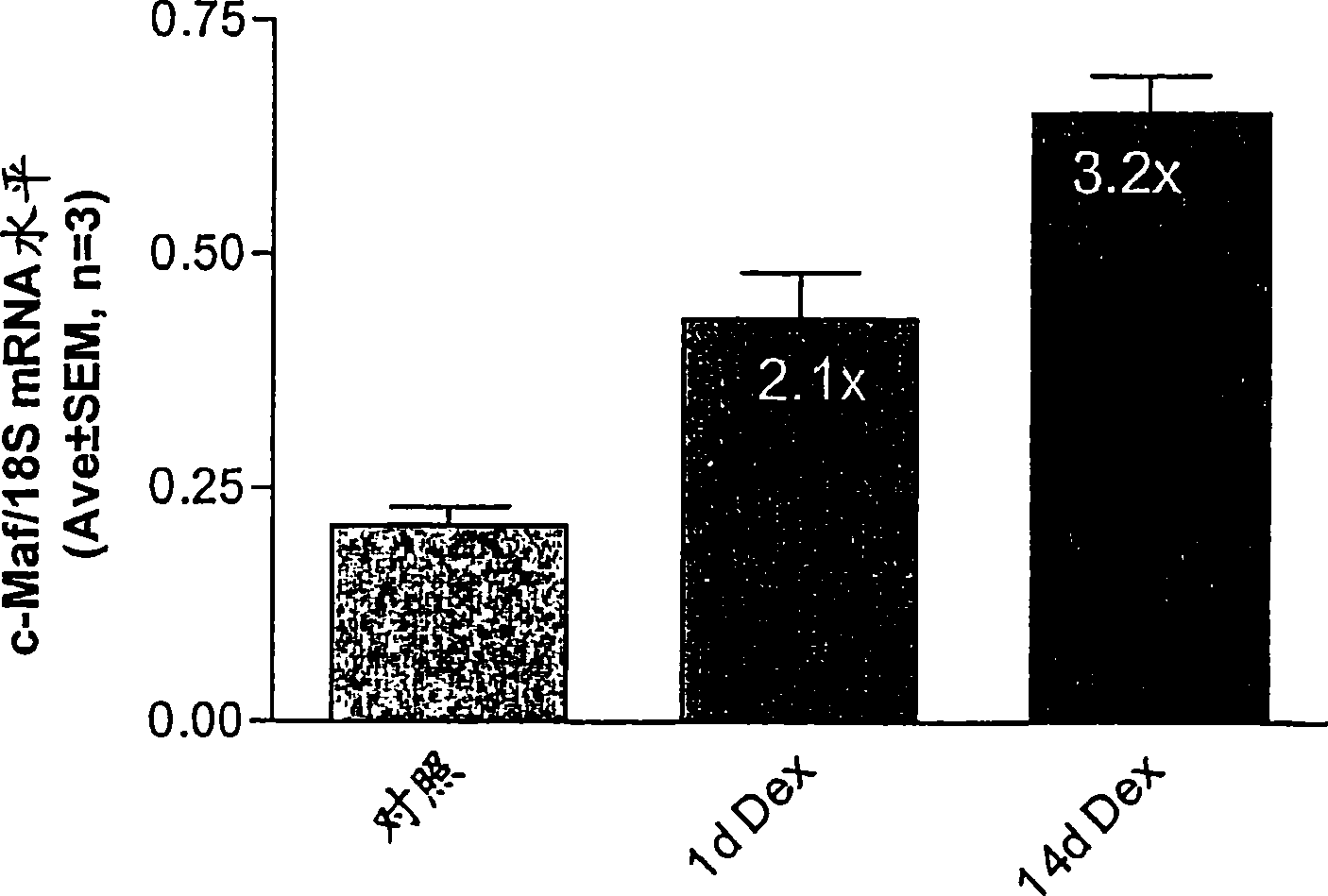CDK2 antagonists as short form C-MAF transcription factor antagonists for treatment of glaucoma
A transcription factor, technology for glaucoma
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Isolation of RNA from Human Trabecular Mesh Tissue and Cells
[0034] Human trabecular meshwork (TM) cells were obtained from donor eyes (Central Florida Lions Eye and Tissue Bank, Tampa, FL) and cultured as previously described (Steely, et al. (1992), Invest Ophthalmol Vis Sci 33(7):2242-50 ; Wilson et al. (1993), Curr Eye Res 12(9): 783-93; Clark, et al. (1994), Invest Ophthalmol Vis Sci 35(1): 281-94.; Dickerson, et al. (1998), Exp Eye Res 66(6):731-8; Wang, et al. (2001), MolVis 7:89-94). TM cells were derived from pools of four normal or glaucomatous cell lines. According to the manufacturer's instructions (Invitrogen, Carlsbad, CA), with TRIZOL Reagents to isolate total RNA from TM cells of each pool.
Embodiment 2
[0036] Affymetrix microarray analysis
[0037] Reverse transcription, second-strand cDNA synthesis, and biotin labeling of amplified RNA were performed according to standard Affymetrix protocols. Human genome U133A and U133B GENECHIPS according to the standard Affymetrix protocol (Affymetrix, Santa Clara, CA) for hybridization, washing and scanning. use GENEARRAY A scanner (Agilent Technologies, Palo Alto, CA) scanned the hybridized GENECHIP array. Raw data were collected and analyzed with Affymetrix Microarray Suite software.
[0038] Filtering of microarray data was performed using GENESPRINCP(R) software (Silicon Genetics, Redwood City, CA). For each experiment, the data for each chip were normalized by dividing each measurement by 50 percent of all signal intensity measurements for that chip. For each experiment, the expression rate for each gene was calculated by dividing the normalized signal for each gene in the treated or diseased samples ...
Embodiment 3
[0040] Quantitative PCR
[0041] First-strand cDNA was generated from 1 μg of total RNA using random hexamers and TAQMAN(R) reverse transcription reagent according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). 100 μl of the reaction was then diluted 20-fold to achieve an effective cDNA concentration of 0.5 ng / μl.
[0042] ABI PRISM was used essentially as described by Shepard, et al. (2001) Invest Ophthalmol Vis Sci 42(13):3173-81 Measurement of short form c-Maf gene expression was performed by quantitative real-time RT-PCR (QPCR) with the 7700 Sequence Detection System (Applied Biosystems). Use PRIMER EXPRESS The software (Applied Biosystems) designed primers for short form specific c-Maf amplification (Genbank accession number AF055376). The forward and reverse primer sequences are TTGGGACTGAATTGCACTAAGATATAA, SEQ ID NO: 1, (nucleotides 3773-3799) and GCGTTCTAAACAGTTTTGCAATTTT, SEQ ID NO: 2, (nucleotides 3823-3847...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com