Molecular stamp sequencing device and sequencing method
A sequencing and detector technology, applied in the field of microarray molecular stamp sequencing devices, can solve the problems of large consumption of drugs, high-throughput detection limitation, and large reaction system, and achieves reduction of raw material consumption, sequencing cost, and dosage. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0021] Example 1. Sequence determination of 50 DNA fragments using rolling circle amplification products.
[0022] 1. Digest the 50 DNA fragments with corresponding restriction endonucleases, and connect one strand of the digested fragments with a universal adapter (containing a 36-base DNA sequence) under the action of the target DNA to form a circular DNA.
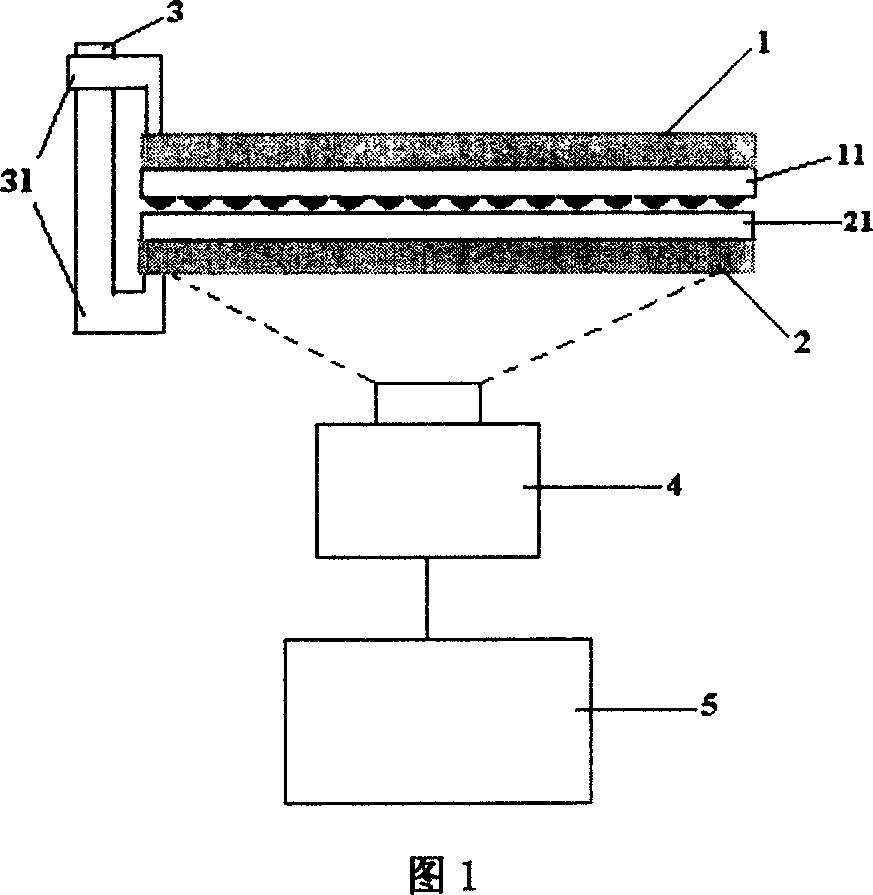
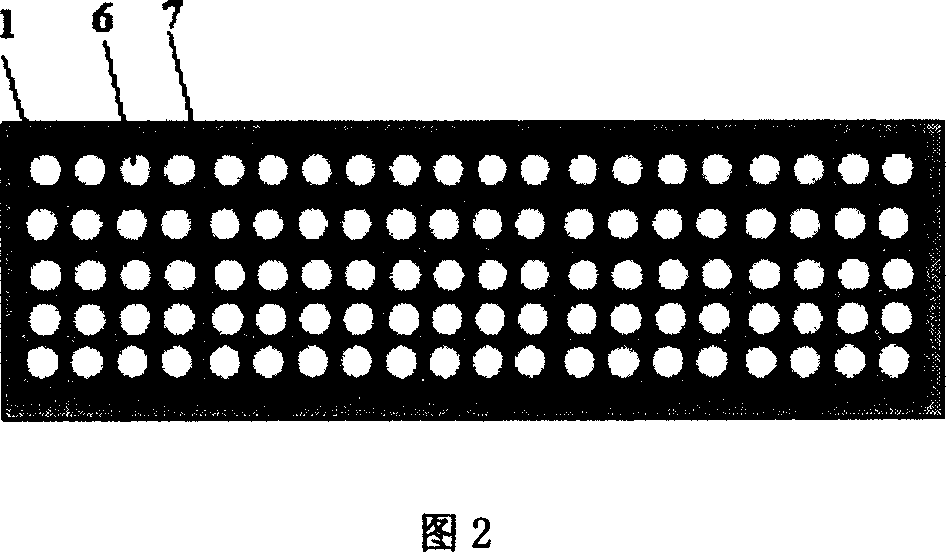
[0023] 2. Use an 18-base universal amplification reverse primer (with acrylamide modification at the 5' end and complementary to the 18 bases at the 3' end of the adapter) and another 18-base universal amplification forward primer (with the adapter 5 ' end 18bps complementary) to the above-mentioned circular DNA by double-primer rolling circle amplification; then the rolling circle amplification products were mixed with acrylamide gel solution, and 50 treated samples were spotted on glass slides with a chip spotter , and aggregated into a gel lattice, and prepared into a microarray chip for sequencing, as shown in Figur...
example 2
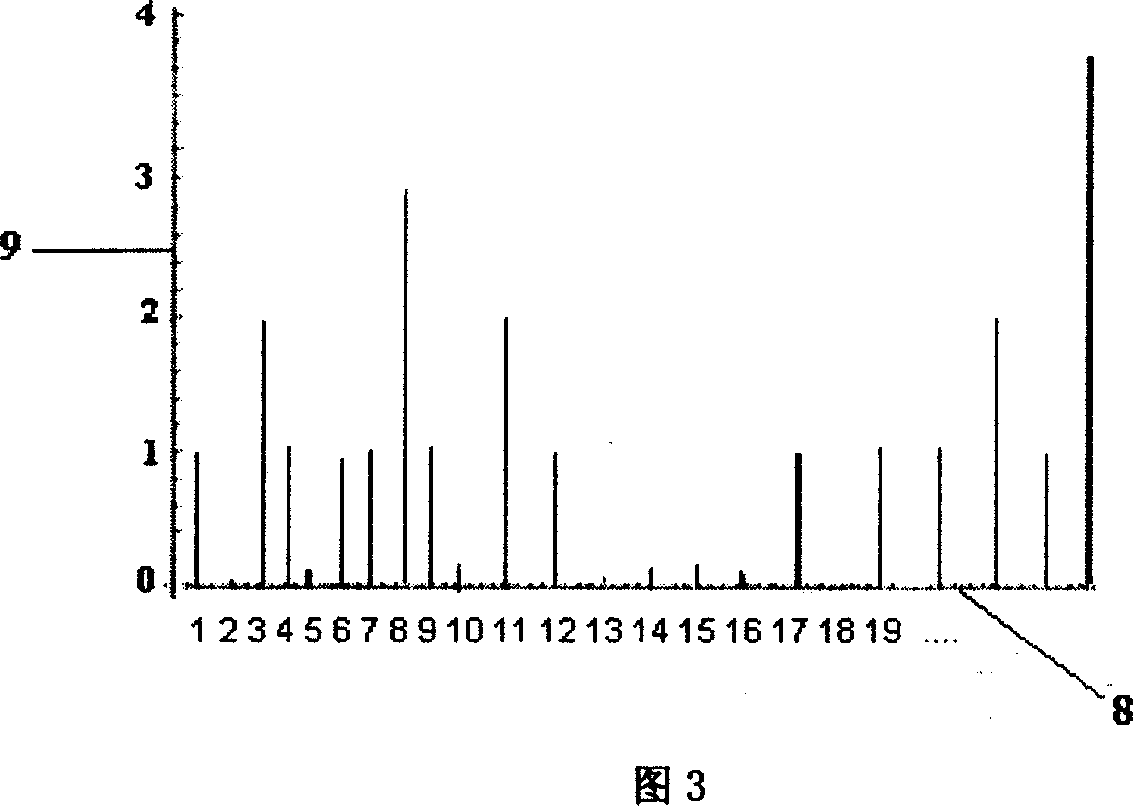
[0030] Example 2. Using PCR products to detect 100 SNP genotypes.
[0031] 1. First, extract human genomic DNA, and obtain 100 fragments of PCR amplification products containing different SNP sites through PCR technology. The 5' end of the reverse primer of the above PCR product is modified with biotin, and the first 18 base sequences of the reverse primer are identical, and the remaining bases are the specific sequence part of each fragment to be amplified.
[0032] 2. The 100 PCR products were respectively fixed with streptavidin-modified magnetic beads, then denatured with NaOH and washed with buffer to obtain single-stranded DNA.
[0033] 3. Fix the above-mentioned single-stranded DNA into a 10×10 array with a magnetic DNA extension substrate.
[0034] 4. Hybridize the microarray DNA on the extension substrate with universal sequencing primers, and wash away excess primers.
[0035] 5. Spread DNA polymerase evenly on the microarray extension substrate of the DNA extensio...
example 3
[0041] Example 3. Re-determination of the whole genome DNA sequence of Escherichia coli using rolling circle amplification products.
[0042] 1. Genomic DNA of Escherichia coli was extracted and fragmented by ultrasonic method. By combining one end of the DNA fragment with a universal 5' terminal phosphate adapter (containing a 36-base DNA sequence) under the action of the target DNA, one strand of the DNA fragment is ligated into a circular DNA, and suitable Dilute the template solution for whole genome sequencing of E. coli to form a single circular DNA.
[0043] 2. Use a 18-base universal amplification reverse primer (with acrylamide modification at the 5' end and complementary to the 18 bases at the 3' end of the adapter) to perform double-primed rolling circle amplification on the above circular DNA; The circle amplification product is mixed with acrylamide gel solution, evenly spread on the glass slide, and polymerized into an ultra-thin gel layer to prepare a rolling c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com