Arrays with modified oligonucleotide and polynucleotide compositions
a technology of oligonucleotide and polynucleotide, which is applied in the direction of biomass after-treatment, organic chemistry, specific use bioreactor/fermenter, etc., can solve the problems of no technique for identifying the type of binding partner, probe sequence on the array, etc., and achieve the effect of speeding up the cycle time for regeneration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis, and Purification of Modified Nucleic Acids
[0124] Oligonucleotides having 2'-5' linkages and 3'-methoxy substitutions were synthesized using commercial phosphoramidites on commercially purchased DNA synthesizers from 1 mM scales using standard phosphoramidite chemistry and methods that are well known in the art, such as, for example, those disclosed in Stec et al., J. Am. Chem. soc. 106:6077-6089 (1984), Stec et al., J. Org. Chem. 50(20):3908-3913 (1985), Stec et al., J. Chromatog. 326:263-280 (1985), LaPlanche et al., Nuc. Acid. Res. 14(22):9081-9093 (1986), and Fasman, Practical Handbook of Biochemistry and Molecular Biology, 1989, CRC Press, Boca Raton, Fla., herein incorporated by reference.
[0125] Oligonucleotides were deprotected following phosphoramidite manufacturer's protocols. Unpurified oligonucleotides were either dried down under vacuum or precipitated and then dried. Sodium salts of oligonucleotides were prepared- using the commercially available DNA-Mate (Bar...
example 2
Specificity of Different Modified Oligonucleotides
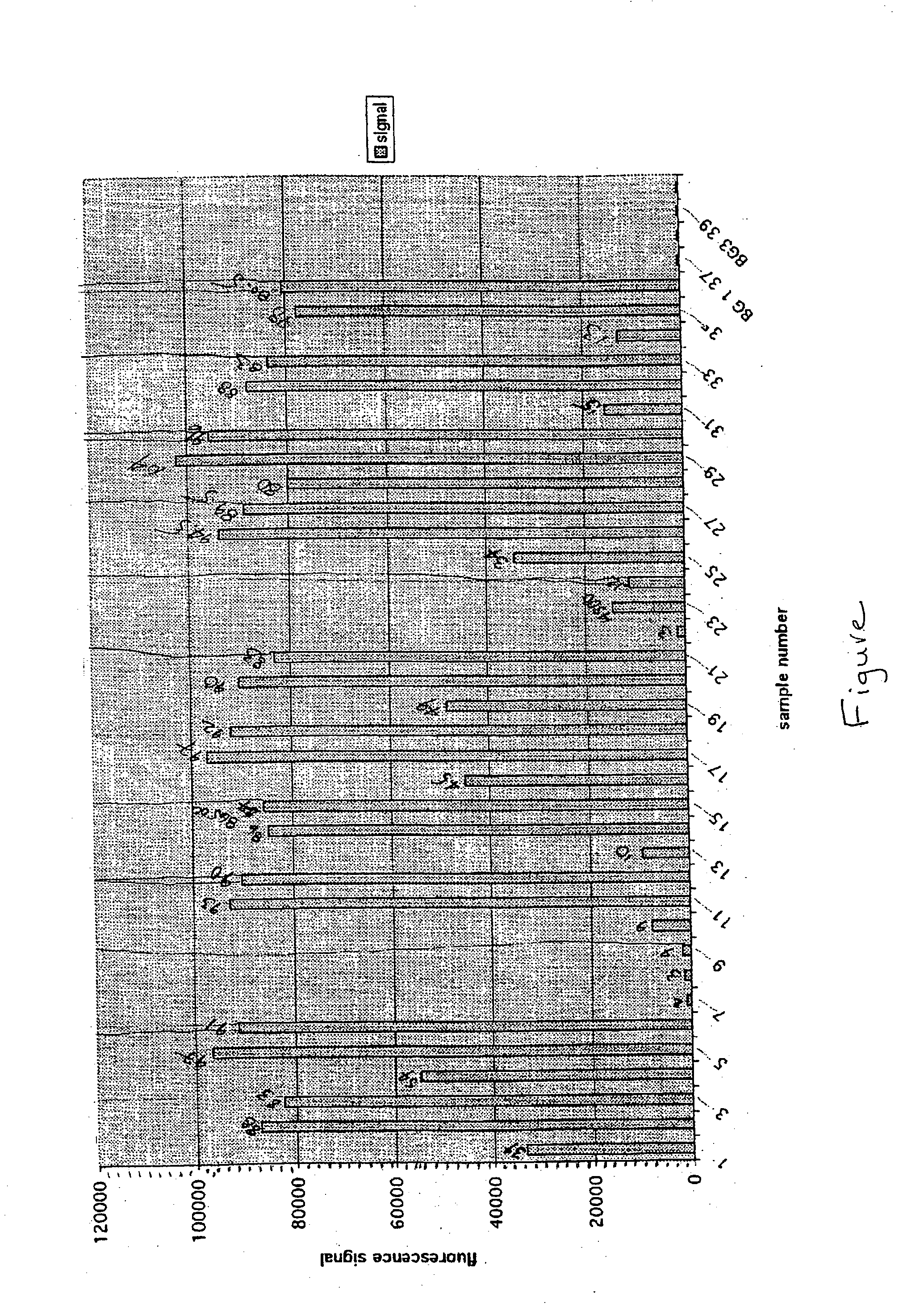
[0131] The binding specificity of 20-mer modified oligonucleotides (5'-ggt ggt tcc tcc tca gtc gg-3'; SEQ ID NO:1) to its RNA or DNA complement (5'-ccg act gag aag gaa cca cc-3'; SEQ ID NO:2) were tested in a solution having 10 mM NaPO.sub.4 with three different NaCl concentrations: no salt, 100 mM NaCl, and 1M NaCl. The RNA or DNA test molecules were labeled with a fluorescent label, Oregon Green 488, to allow detection of these molecules upon hybridization to the test sequences. Following hybridization, the level of binding of each of these molecules was determined by the level of Oregon Green 488 that was detectable on the controlled pore glass (CPG) beads having the specific modified oligonucleotides.
[0132] Results were as follows:
1TABLE 1 Binding of Polymers Having Different Backbones with RNA and DNA Sample numbers are shown in parenthesis. Binding Partners No salt 100 mM NaCl 1000 mM NaCl 2'-O-Me DNA (1) 133207 (2) 86762 (3) 8...
example 3
Isoation and Detection using a Bead Array
[0134] Magnetic beads are coated with 2'-5'linked 3'-methoxy substituted oligonucleotides having sequences complementary to transcripts of genes known to be involved in breast cancer, such as HER-2, p53, ras and the like. Each bead is coated with multiple copies of an oligonucleotide having the same gene-specific sequence. Approximately 100 ng of each oligonucleotide are added to 1 .mu.g of coated magnetic beads and incubated for 10 minutes. Magnetic beads were collected using a cobalt magnet and washed in PBS, followed by resuspension in 1 ml PBS, and the beads combined for the creation of the bead array. All incubations are performed with gentle agitation or vortex mixing at room temperature.
[0135] Cells taken from a breast tumor are assayed by exposing 1 ml samples of homogenized tumor tissue and 1 ml of homogenized normal breast tissue, diluted 1:10 in PBS, with 1 to 2 .mu.g of the bead array for 1 hour at room temperature and washed thre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com