Fed-batch fermentation process and culture medium for the production of plasmid DNA in E. coli on a manufacturing scale
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Fed-Batch Fermentation of E. coli JM108 Carrying the Plasmid pRZ-hMCP1
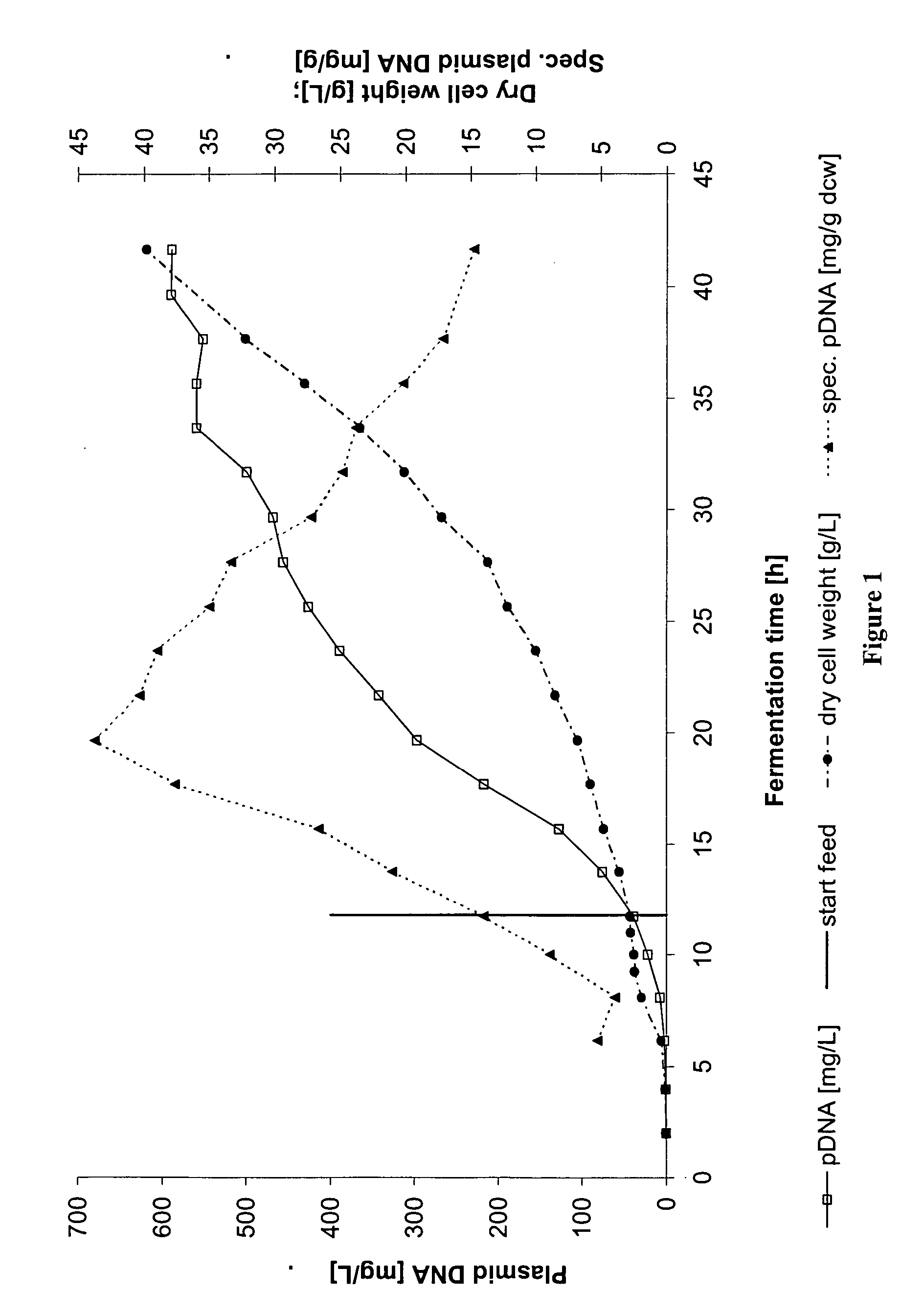
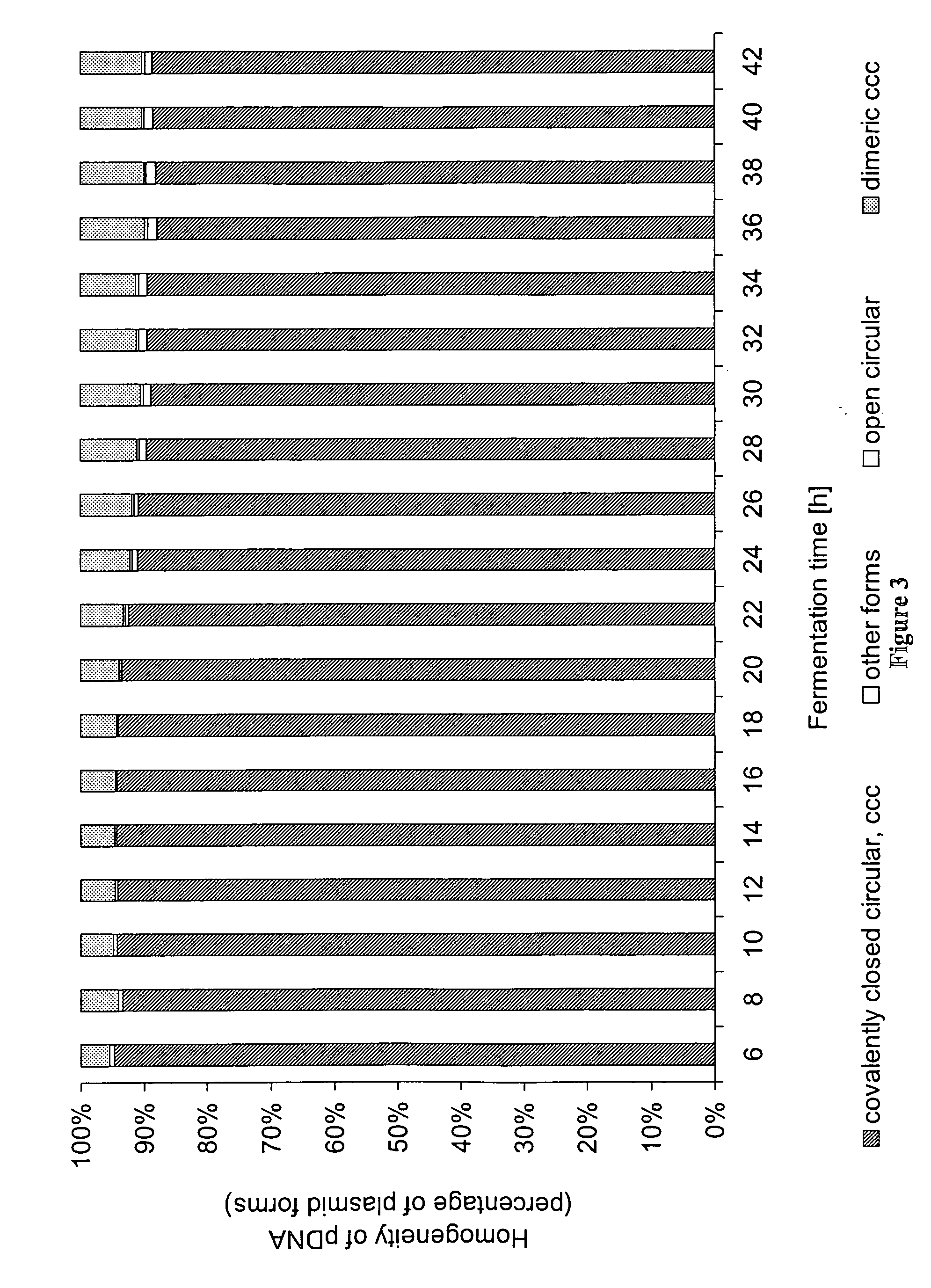
[0113] An exponential fed-batch fermentation was carried out in a 20 L scale fermenter (stirred tank reactor) with the E. coli K-12 strain JM108 harboring the plasmid pRZ-hMCP1. This plasmid (4.9 kb) is a derivative of pcDNA3™ (Invitrogen) containing a pUC ori and a kanamycin resistance marker. The gene of interest of pRZ-hMCP1 is monocyte chemoattractant protein 1 (Furutani et al., 1989) under the transcription control of the eukaryotic CMV (cytomegalo virus) promoter.
[0114] For a pre-culture, a glycerol stock of the strain (300 μL) was inoculated into a baffled 1000 mL shake flask containing 300 mL of a defined medium. This was cultivated in a rotary shaker at 300 rpm and 37° C. The pre-culture medium was composed as follows: NH4Cl 2 g / L, MgSO4*7H2O 0.24 g / L, glucose 10 g / L, L-proline 0.2 g / L, L-isoleucine 0.2 g / L, thiamine hydrochloride 1 mg / L, citric acid 2 g / L, KH2PO4 5.44 g / L, Na2HPO4*12H2O 14.38 g / L and ...
example 2
Influence of Isoleucine on the Plasmid Yield in an Exponential Fed-Batch Fermentation of E. coli JM108
[0123] In 1 L scale screening fermenters, the effect of isoleucine on growth and plasmid production was shown by applying the process described in Example 1.
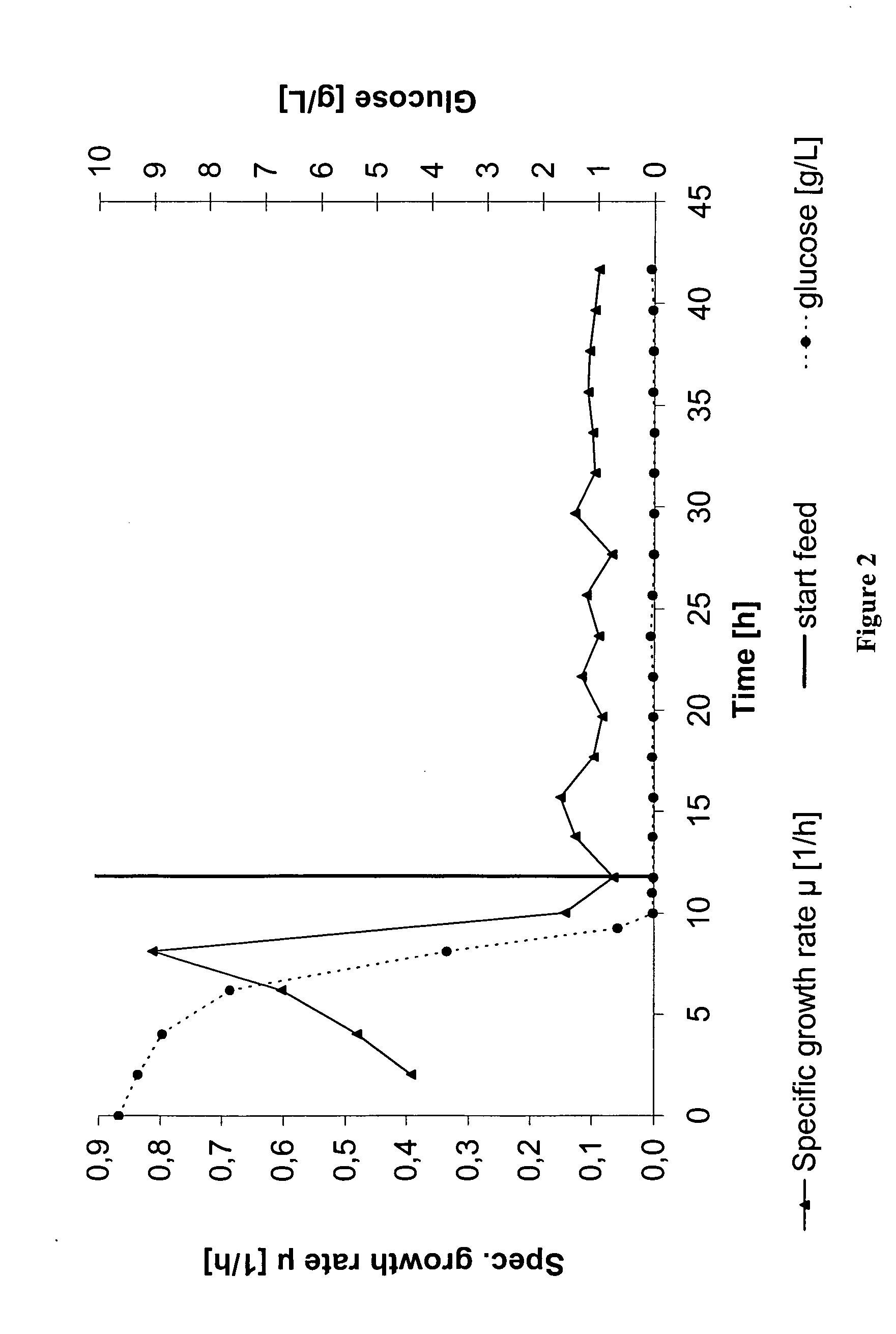
[0124] Two fermentations were carried out in the same way as described in Example 1, with the only difference that one medium contained isoleucine whereas the other medium did not. The remaining composition of the culture medium was identical as described in Example 1 as well as the cultivation conditions and the mode of exponential feeding at the growth rate of p=0.1 h−1. The feeding rate was automatically controlled via a balance and peristaltic pumps.
[0125] The time course of the optical density and the volumetric pDNA yield of both fermentations is shown in FIG. 5 (with isoleucine) and FIG. 6 (without isoleucine). Growth of both fermentations was nearly identical with an average specific growth rate of p=0.09 h−1 during ...
example 3
Fed-Batch Fermentation of E. coli JM108 Carrying the Plasmid pRZ-hMCP1 (20 L Fermenter), using an Exponential Feeding Algorithm, Succeeded by a Linear Feeding Mode
[0127] In this Example, E. coli JM108 cells carrying the plasmid pRZ-hMCP1 are prepared and cultivated as described in Example 1. Other than in Example 1, the feeding phase is divided into two different parts: [0128] (1) an exponential feeding phase, where the feeding rate follows an exponential feeding function in order to maintain a specific growth rate of μ=0.25 h−1, and [0129] (2) a linear constant feeding phase, where the feeding rate is maintained at a constant value of 200 mL / h.
[0130] The time point, when switching from exponential to linear feeding takes place, is after 10 hours of exponential feeding. The linear feeding phase is chosen to be 10 hours. By such fermentation, volumetric and specific plasmid yields are obtained that range from 500 to 800 mg pDNA / L or 20 to 30 mg pDNA / g DCW.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Density | aaaaa | aaaaa |
| Density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com