Function homology screening
a screening and function technology, applied in the field of function homology screening, can solve the problems of affecting the accuracy of the detection results, so as to enhance the response of the measured components and assess the effect of a perturbation on gene expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Regulators of Endothelial Cell Responses to Inflammation
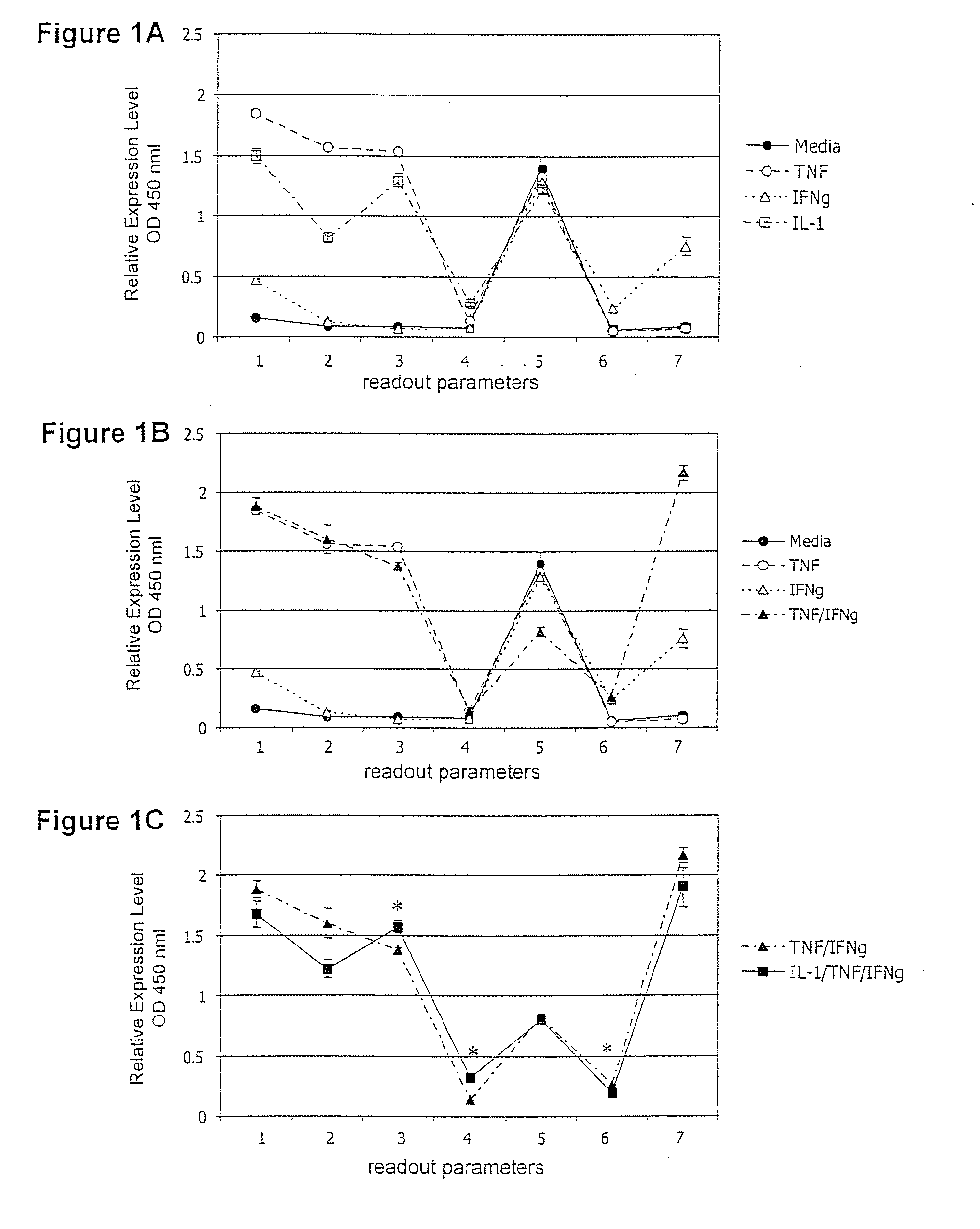
[0177] The present invention is useful for identifying regulators of inflammation using human endothelial cells as an indicator cell type. A set of assay combinations that reproduces aspects of the response of the endothelial cells to different types of inflammatory processes is developed in vitro.
[0178] Primary human umbilical vein endothelial cells (HUVEC) are used. Other cells that may replace HUVEC in the screen include primary microvascular endothelial cells, aortic or arteriolar endothelial cells or endothelial cell lines such as EAhy926 or E6-E7 4-5-2G cells or human telomerase reverse transcriptase-expressing endothelial cells (Simmons, J. Immunol., 148:267, 1992; Rhim, Carcinogenesis 19:673, 1998; Yang, J. Biol. Chem. 274:26141, 1999). 2×104 cells / ml are cultured to confluence in EGM-2 (Clonetics). Other media that may replace EGM-2 include EGM (Clonetics) and Ham's F12K medium supplemented with 0.1 mg / ml heparin and...
example 2
Multiplex Assay Combinations for Distinguishing Mechanism of Action
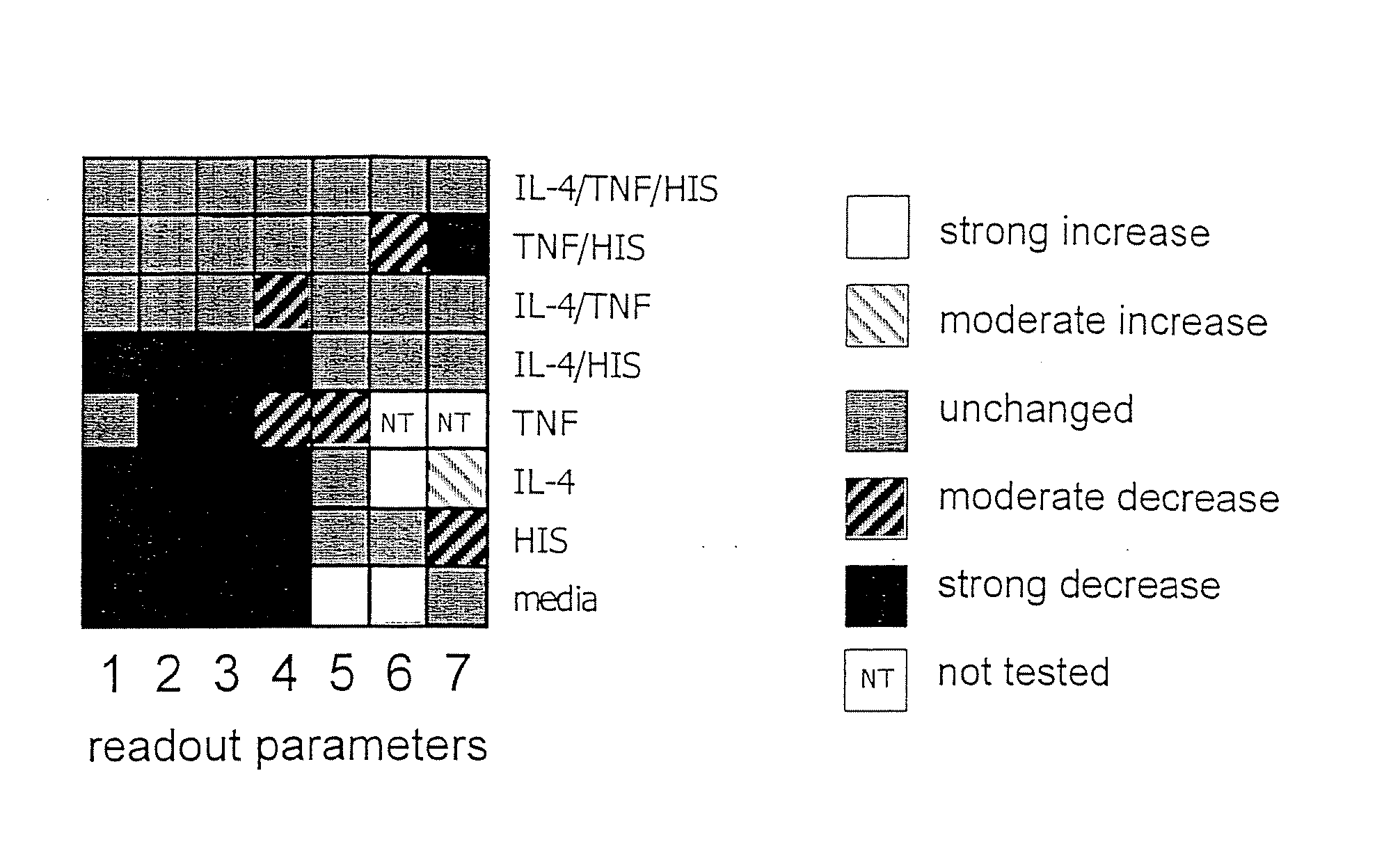
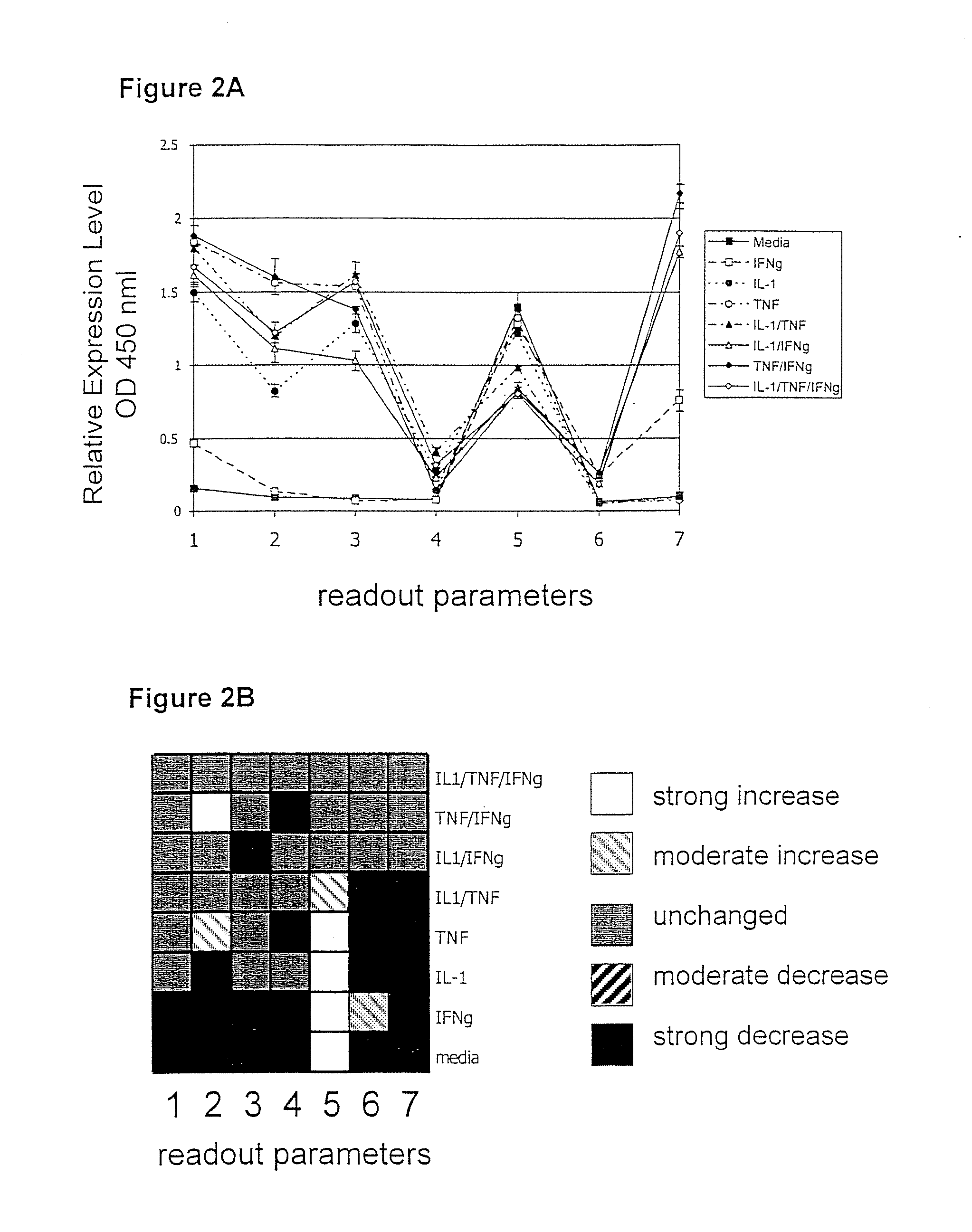
[0204] The following example demonstrates the utility of the invention in identification of the mechanism of action of a test compound or intervention identified in the optimized assay combination of Example 1. This assay combination is included in a panel that contains specific and targeted alterations. A neutralizing antibody to TNF-α was selected as a test agent, as it is active when tested in the optimized primary assay combination of Example 1 (FIG. 3A). When the test agent is evaluated in the panel of assay combinations, it can be determined if the active compound is acting on a component(s) unique to one receptor-stimulated pathway, or on a common pathway component or pathway activity. The neutralizing antibody to TNF-α as a test agent evaluated in these assay combinations alters the biomap, as shown in FIG. 3B.
[0205] Confluent cultures of HUVEC cells are treated with TNF-α (5 ng / ml), IFN-γ (100 ng / ml), IL-1...
example 3
Analysis in Multiplex Assay Combinations for Identifying Mechanism of Action
[0209] The following example demonstrates the usefulness of the present invention for identification of mechanism of action of a test agent selected as an active agent. A recombinant fusion protein containing the extracellular domains of the p55 TNF-α-receptor fused to immunoglobulin Fc domain (p55-Fc fusion protein) is selected as an active compound when tested in the optimized assay combination of Example 1 (FIG. 7A).
[0210] Confluent cultures of HUVEC cells are treated with TNF-α (5 ng / ml)+IFN-γ (100 ng / ml)+IL-1 (20 ng / ml) in the presence or absence of p55-Fc (50 ng / ml). After 24 hours, cultures are washed and evaluated for the cell surface expression of ICAM-1 (1), VCAM-1 (2), E-selectin (3), CD31 (4), and MIG (5) by cell-based ELISA performed as described in FIG. 1. The relative expression of each parameter is shown in FIG. 7A along the y-axis as average value of the OD measured at 450 nm of triplicate...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com