Method for determining base sequence of DNA
a technology of dna and base sequence, applied in the field of determining the base sequence of dna, can solve the problems of enormous cost and effort, and achieve the effect of short tim
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Incorporation of a Sequence-Specific Fluorescent Probe in a Multimolecular System
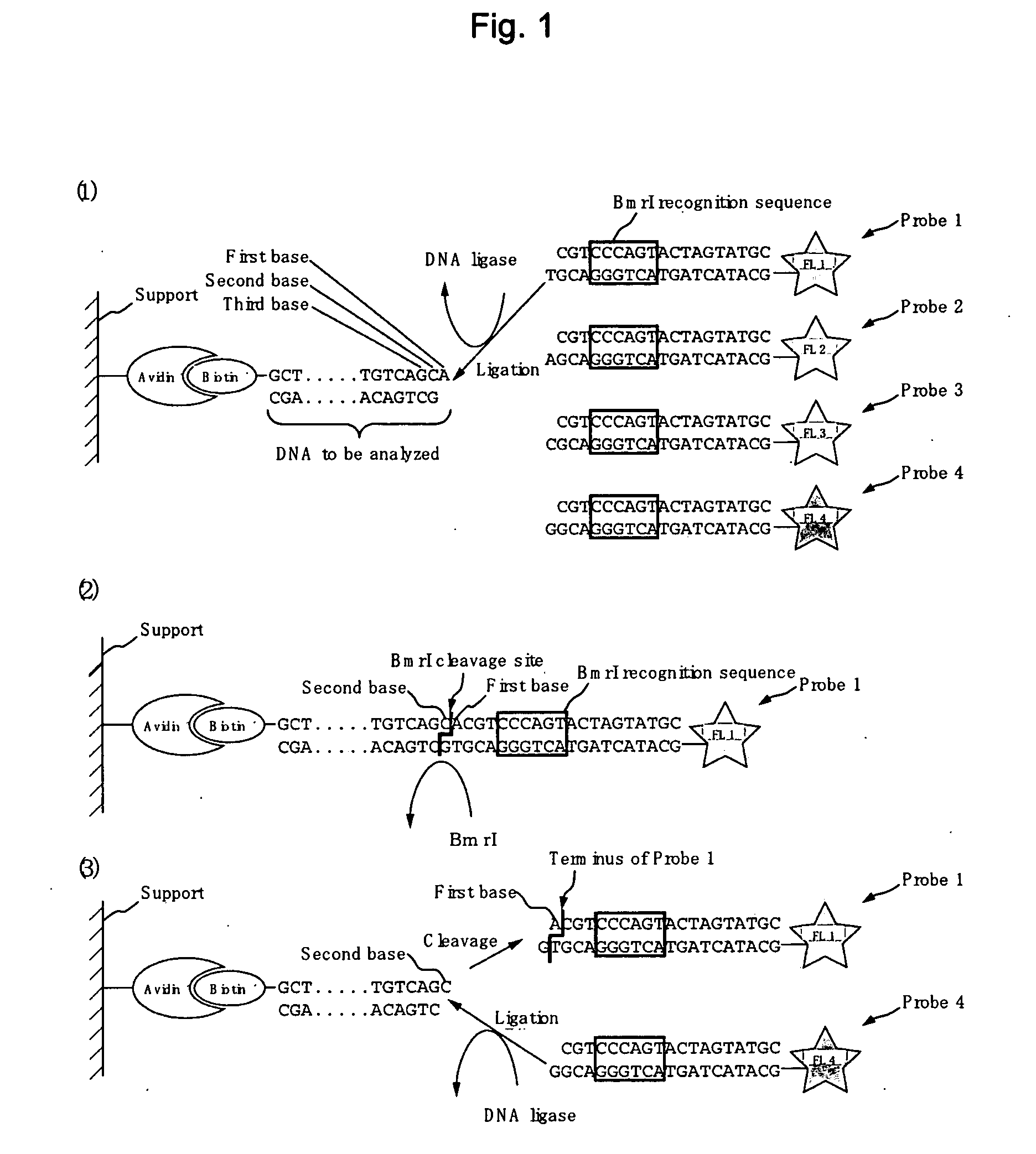
[0133]The base sequence of a DNA was determined by detecting a fluorescent DNA probe that binds specifically to the sequence of a DNA having a one base-protruding end and further reacting alternately a Class IIS restriction enzyme and a DNA ligase.
(1) Immobilization of DNA to be Analyzed onto a Support
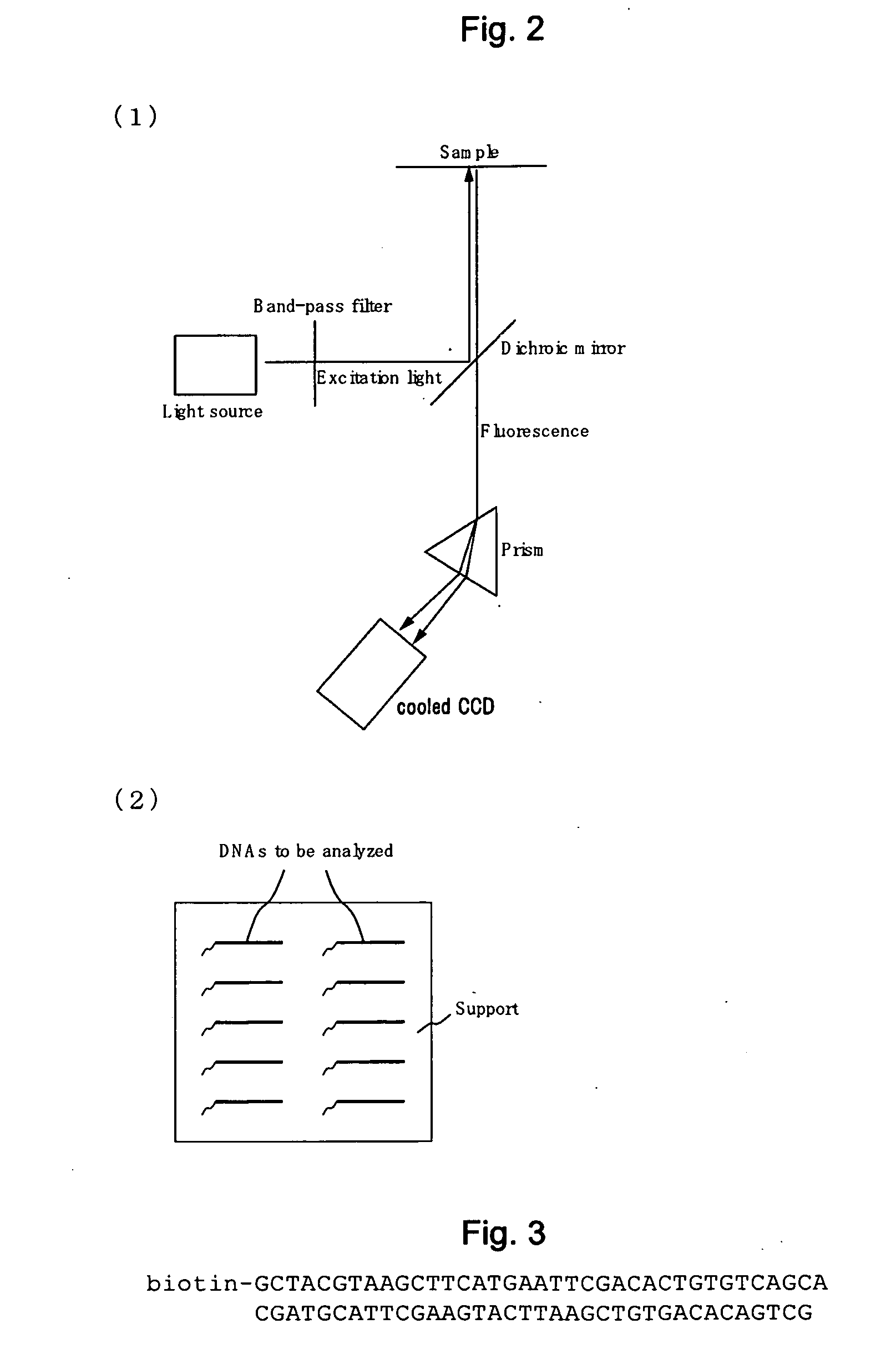
[0134]Biotinylated DNA comprising biotin conjugated to the DNA to be analyzed (FIG. 3) (SEQ ID No.: 9: 5′ biotin-GCTACGTAAGCTTCATGAATTCGACACTGTGTCAGCA 3′, SEQ ID No.: 10: 5′ GCTGACACAGTGTCGAATTCATGAAGCTTACGTAGC 3′) (10 μM) was mixed with avidin-agarose beads (Sigma, A9207) (suspended with 1×PBS into a 30 μl slurry) and admixed for one hour to immobilize the DNA to be analyzed onto avidin-agarose beads.
(2) Ligation of the DNA to be Analyzed and Fluorescent DNA Probes
[0135]Beads having the DNA to be analyzed obtained immobilized in (1) above were washed twice with 1×PBS, further washed once with a T4 DNA lig...
example 2
[0144]A combination of base sequences of DNA was analyzed by single molecule observation of a chain reaction by a restriction enzyme and a ligase.
(1) Preparation of Quantum Dot-DNA Probes
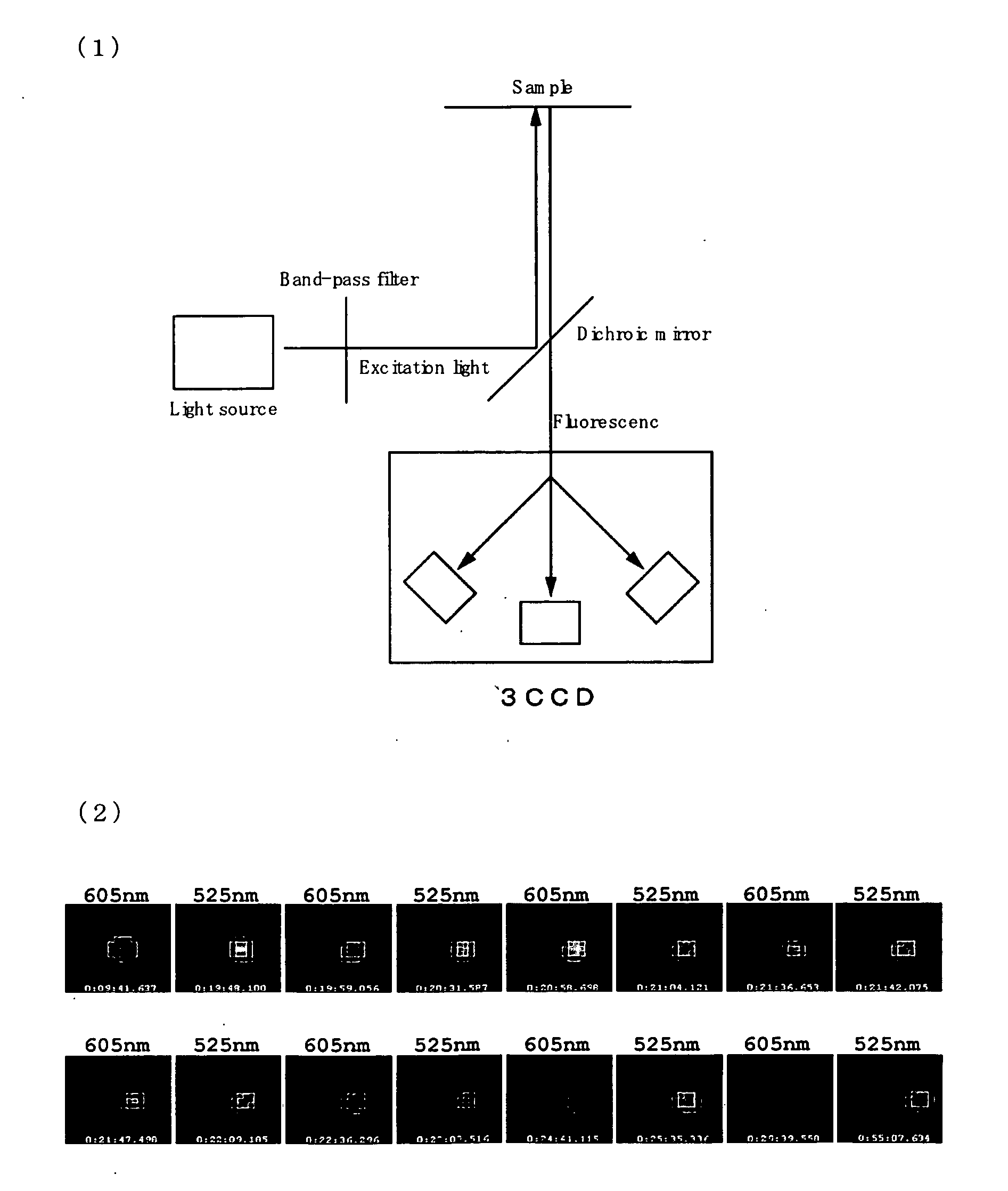
[0145]Using a crosslinking agent (EDC), quantum dots (525 and 605) (Q21341MP Qdot (registered trade mark) 525 ITK™ Carboxyl Quantum Dots, and Q21301MP Qdot (registered trade mark) 605 ITK™ Carboxyl Quantum Dots, Invitrogen Corporation) were respectively bound covalently to the amino groups at 5′ of the DNAs described below to prepare quantum dot-DNA probes.
Quantum dot 525 (FIG. 5 (1a)):SEQ ID No.: 11: 5′ cgtcccagtactagtatgc 3′SEQ ID No.: 12: 5′ amine-gcatactagtactgggacgc 3′Quantum dot 605 (FIG. 5 (1b)):SEQ ID No.: 13: 5′ cgtcccagtactagtatgc 3′SEQ ID No.: 14: 5′ amine-gcatactagtactgggacgt 3′
(2) Immobilization of the DNAs to be Analyzed onto a Support
[0146]First, avidin (Sigma, A9275) was immobilized on the surface of a cover glass. Biotinylated DNA comprising biotin bonded to the DNA to be analyzed (...
example 3
[0149]The base sequence of a DNA was identified by way of single molecule observation of a chain reaction by a restriction enzyme and a ligase, using a support with a plurality of DNAs to be analyzed immobilized.
(1) Phosphorylation of Oligo DNA
[0150]Added with 22 μl of 10× T4 polynucleotide kinase buffer (NEB), 5 μl of ATP (100 mM) and 10 μl of T4 polynucleotide kinase (NEB), 200 μl of oligo DNA (50 μmol / μl) (Hokkaido System Science) was incubated at 37° C. for five hours. After incubation, phenol extraction (phenol: chloroform=1:1, stirring with a Vortex mixer for 10 seconds), chloroform wash (stirring with a Vortex mixer for 10 seconds), isopropanol precipitation (treatment for one minute with 50% isopropanol and 0.3M acetic acid sodium at room temperature, then centrifugation at 20000×g for 15 minute at 4° C.), and 70% ethanol wash (treatment with 1000 μl, then, centrifugation at 20000×g; number of washes: once) were carried out. After washing, [the oligo DNA] was dissolved in 10...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| distance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com