Compositions and methods for delivering nucleic acid to a cell
a nucleic acid and cell technology, applied in the field of compositions and methods for delivering nucleic acid to cells, can solve the problems of poor in vivo transfection efficiency, insufficient stability of liposome-nucleic acid complexes, and inability to commercialize therapeutic nucleic acid containing liposomes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Lipid Formulation with Added Polyamine
Abbreviations
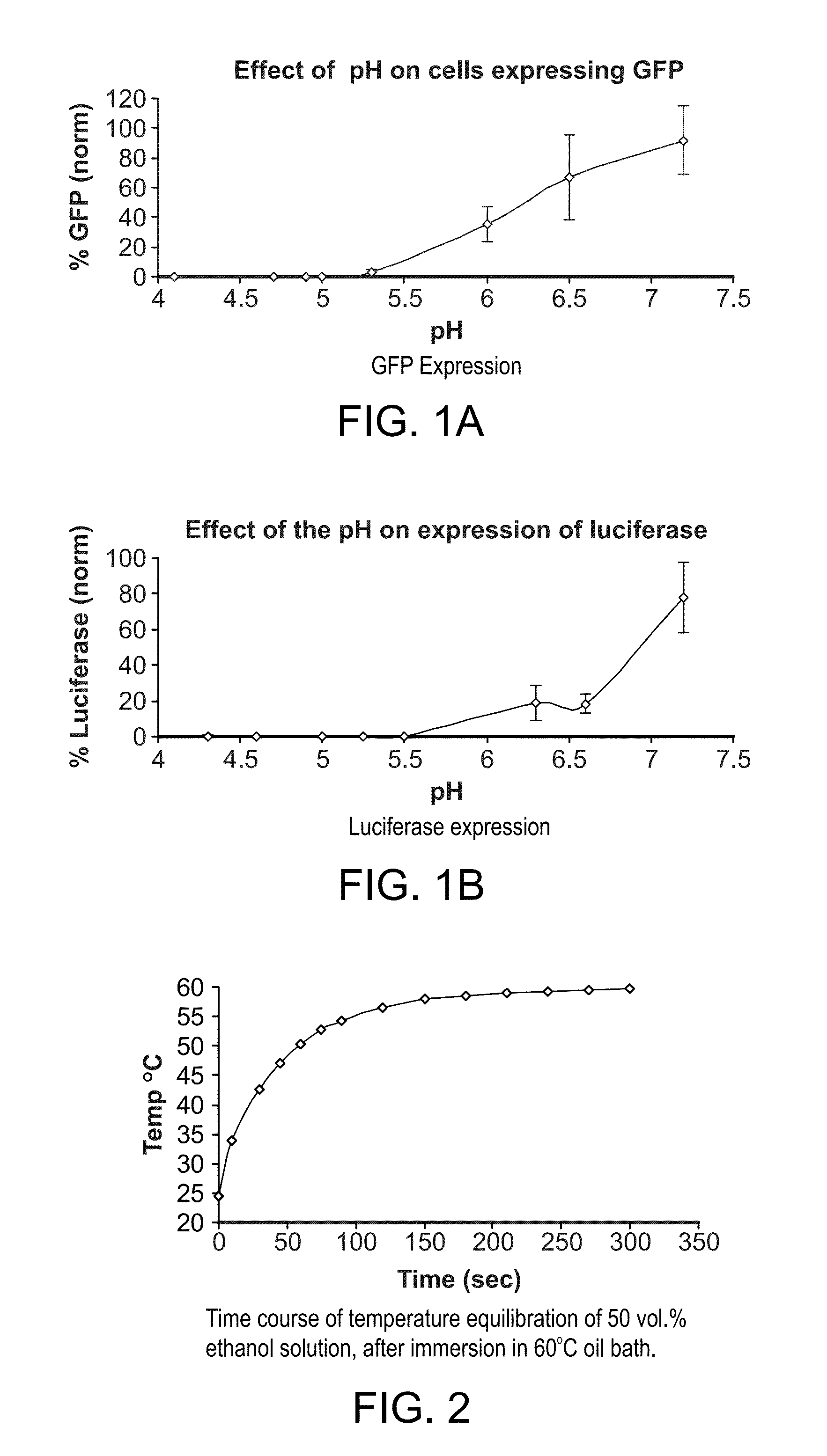
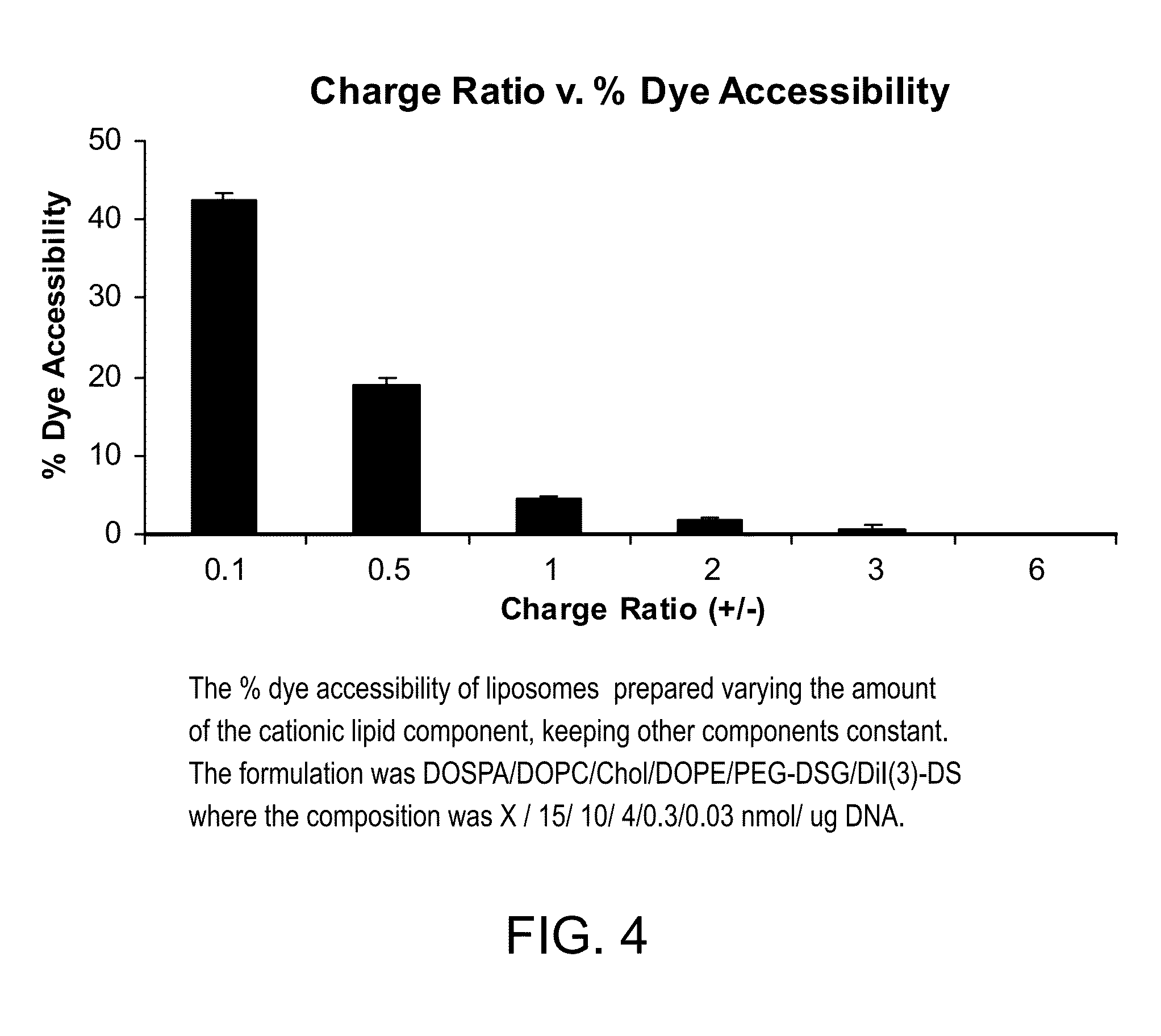
[0085]GFP: Green fluorescent protein[0086]MES: 2-(N-morpholino)ethanesulfonic acid[0087]TE buffer: Tris / EDTA buffer[0088]HBS: HEPES buffer[0089]PEI: polyethyleneimine[0090]Hank's' BSS: Hank's' balanced salt solution[0091]DOTAP: dioleoyl trimethylammonium propane[0092]DOPC: 1,2-dioleoyl-sn-glycero-3-phosphocholine[0093]Chol: cholesterol[0094]DOPE: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine[0095]CHEMS: cholesteryl hemisuccinate[0096]PEG: polyethylene glycol[0097]PEG-DSG: PEG-distearoylglycerol[0098]DOSPA: N-(1-(2,3-dioleyloxy)propyl)-N-(2-(sperminecarboxamido)ethyl)-N,N-dimethyl-ammonium trifluoroacetate[0099]CHIM: cholesterol imidazole derivative[0100]DiI(3)-DS: a cationic lipid dye:
Liposome Preparation
[0101]The polyamine was mixed with GFP plasmid DNA in 50% ethanol / 50% 20 mM MES, pH 5.1. The lipid mixtures were dissolved in 50% ethanol / 50% 2 mM TE buffer, pH 8.0. The polyamine mixture and the lipid mixture were heated at 60° C. ...
example 2
Method
[0110]DNA (100 μg) was prepared in a 50% ethanol solution as described above. From a stock solution of PEI 600, aliquots were added to in such volumes to give N / P=0, 0.67, 1.33 and 4. Separately, a solution of 2 mM TE buffer, pH 8.0 and ethanol (50% v / v) were prepared. The solution were heated at 60° C. for 2 min and mixed. The samples were cooled and dialysed as above. DNA concentration and dye accessibility was measured as above.
Results
[0111]
%plate1plate2DyeSample[dna]ng / mlstdev[dna]ng / mlstdevAccesstdev[DNA]ug / mlstdevnakedDNA729.116.8779.1217.893.62.3123.370.53N / P 0.67695.121.76771.9617.9590.02.1123.160.54N / P 1.33505.753.72527.7919.2395.83.5615.830.58N / P 4.0467.522.63506.8517.892.23.2815.210.53
Conclusion
[0112]Addition of PEI to DNA does not inhibit the binding of Picogreen®. Therefore any protection afforded to DNA during the liposome encapsulation method even with PEI included within the formulation must come from lipid encapsulation.
example 3
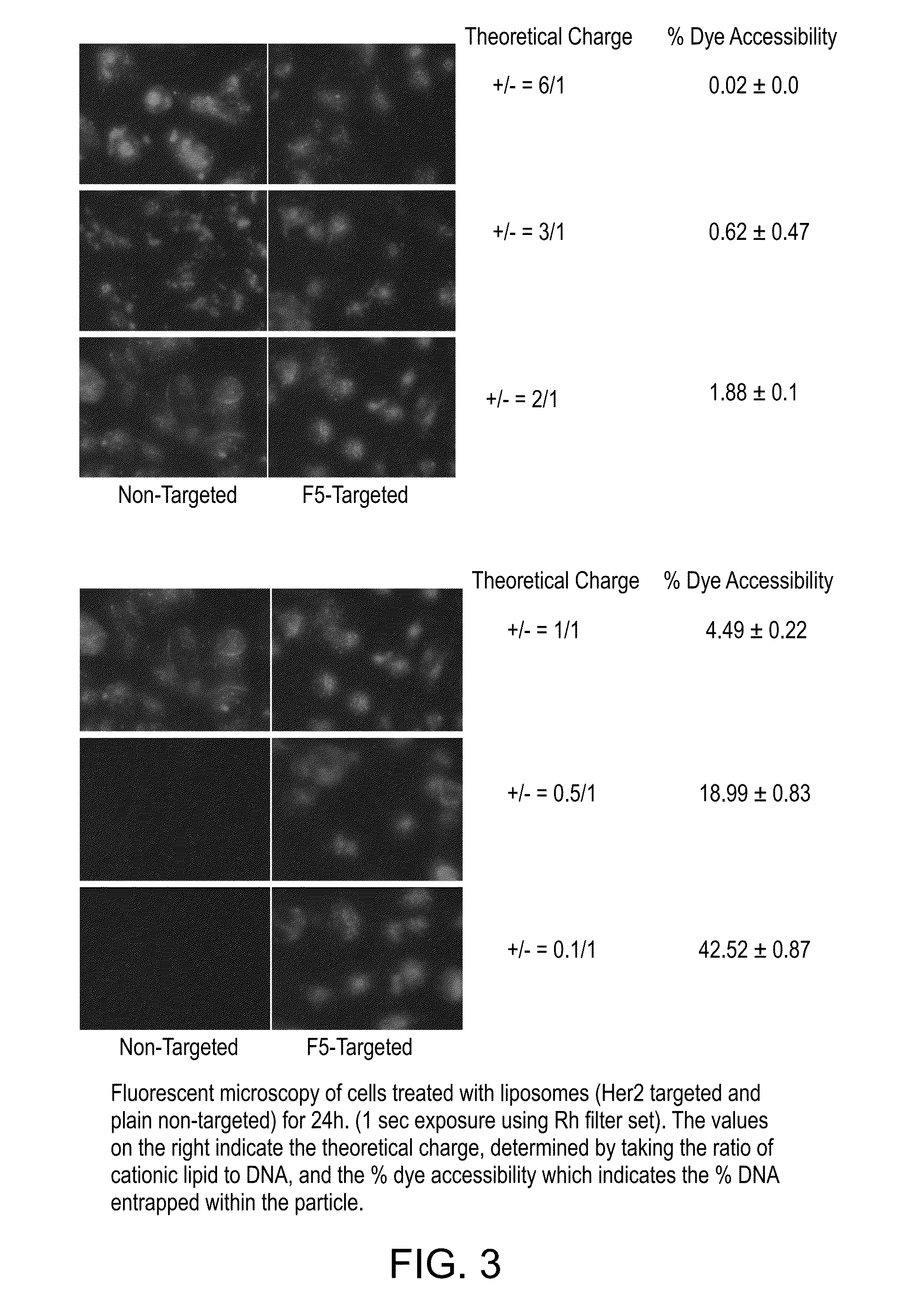
[0113]The following experiment was conducted to investigate whether cationic lipid can be removed from the liposome composition.
Method
[0114]Lipid Formulation=DOTAP / DOPC / Chol / PEG-DSG / DiI(3)-DS 30 / 1500 / 1000 / 30 / 3 nmol per μg DNA
Formulation 1: Lipid as above mixed with DNA (plus PEI N / P=0.9)
Formulation 2: DNA plus PEI N / P=0.9
Formulation 3: Lipid as above mixed with DNA
Formulation 4: Lipid as above (except no DOTAP) mixed with DNA (plus PEI N / P=0.9)
Results
[0115]
% DyeFormulationAccessStdevSize nm121.50.9183.5 ± 60293.22.4N.D331.21248.8 ± 67.2424.00.7289.6 ± 112.5
Conclusion
[0116]Using the DNA pre-contacted with a polyamine or polymeric amine, liposomal particles can be made without using any cationic lipid, that entrap >75% of the DNA.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Substance count | aaaaa | aaaaa |
| Substance count | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com