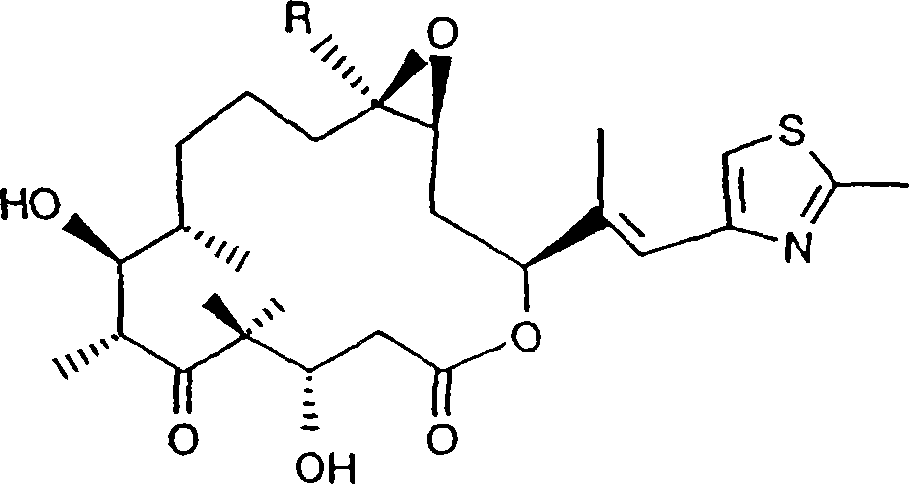
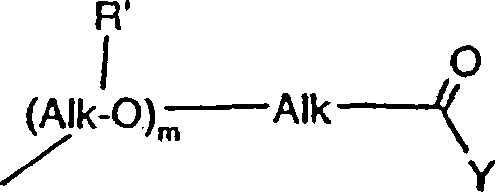Genes for the biosynthesis of epothilones
A biosynthetic, 0.5mnapo4ph7.0 technology, used in plant genetic improvement, genetic engineering, biochemical equipment and methods, etc., can solve the problems of complex genes and unisolated
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
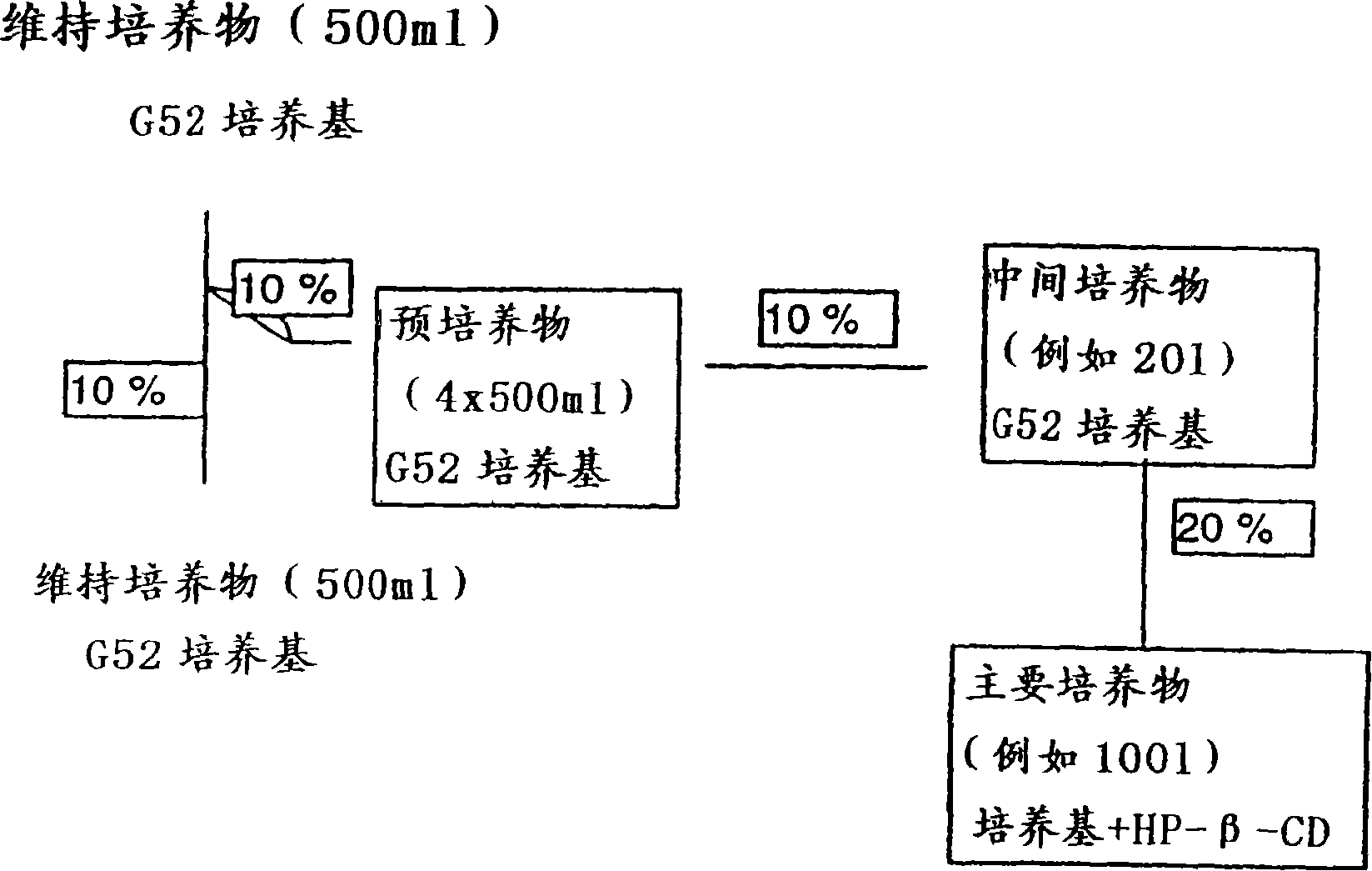
[0114] Example 1: Cultivation of S. cellulosus strains producing epothilone
[0115] Cellulose bacteria strain 90 (DSM 6773, Deutsche Sammlung vonMikroorganismen und Zellkulturen, Braunschweig) in SolE medium (0.35% glucose, 0.05% tryptone, 0.15% MgSO 4 ·7H 2 O, 0.05% ammonium sulfate, 0.1% CaCl 2 , 0.006%K 2 HPO 4 , 0.01% sodium dithionite, 0.0008% Fe-EDTA, 1.2% HEPES, 3.5% [vol / vo / ] sterilized supernatant of a culture of S. cellulosus in stationary phase) pH adjusted to 7.4 on an agar plate Streak and incubate at 30°C. Pick from 1cm 2 The cells were inoculated into 5ml G51t liquid medium (0.2% glucose, 0.5% starch, 0.2% tryptone, 0.1% probion S, 0.05% CaCl 2 2H 2 O, 0.05% MgSO 4 ·7H 2 O, 1.2% HEPES, pH adjusted to 7.4), and cultured at 30° C. with shaking at 225 rpm. After 4 days, the culture was transferred to 50 ml G51t and grown for 5 days as described above. This culture was used to inoculate 500 ml of G51t and cultured for 6 days as described above. The cult...
Embodiment 2
[0116] Example 2: Construction of Bacterial Artificial Chromosome Library
[0117] To construct the Bac library, S. cellulosus cells cultured as described in Example 1 were embedded in agarose blocks, lysed, and the released genomic DNA was partially digested with the restriction enzyme HindIII. Digested DNA was separated by pulse electrophoresis on an agarose gel. Large fragments of DNA (approximately 90-150 kb) were isolated from agarose gels and ligated into the vector pBelobacII. pBelobacII contains the gene encoding chloramphenicol resistance, a multiple cloning site within the lacZ gene providing blue-white selection on appropriate media, and genes required for replication and maintenance of one or two copies of the plasmid per cell. Using conventional electroporation techniques, the ligation mixture was transformed into E. coli DH10B electrocompetent cells. Chloramphenicol-resistant recombinant (white spot, lacZ mutation) colonies were transferred to positively charge...
Embodiment 3
[0118] Example 3: Screening of related sequences of the first class of polyketide synthases in the Bac library of S. cellulosus 90
[0119] Bac library filters were probed using a conventional Southern hybridization procedure. The probes used encode the beta-ketoacyl synthase domains of the first and second modules of the rifamycin polyketide synthase (Schupp et al., FEMS Microbiology Letters 159:210-207 (1998)). Probe DNA was generated by PCR using primers flanking each ketosynthase domain using plasmid pNE95 (pNE95 is cosmid 2 described in Schupp et al. (1998)) as a template. 25 ng of PCR-amplified DNA was isolated from a 0.5% agarose gel and labeled with a random primer kit (Gibco-BRL, Methesda, MD, USA) according to the supplier's instructions. 32 P-dCTP labeling. Hybridization was performed at 65°C for 36 hours, and the membrane was washed under highly stringent conditions (0.1xSSC and 0.5% SDS at 65°C for 20 minutes three times). The labeled spots were exposed to a phos...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com