Process for production of succinic acid
一种琥珀酸、核苷酸的技术,应用在基于微生物的方法、生物化学设备和方法、微生物等方向,能够解决不知道琥珀酸合成途径、尚未阐明基因实际功能等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097]
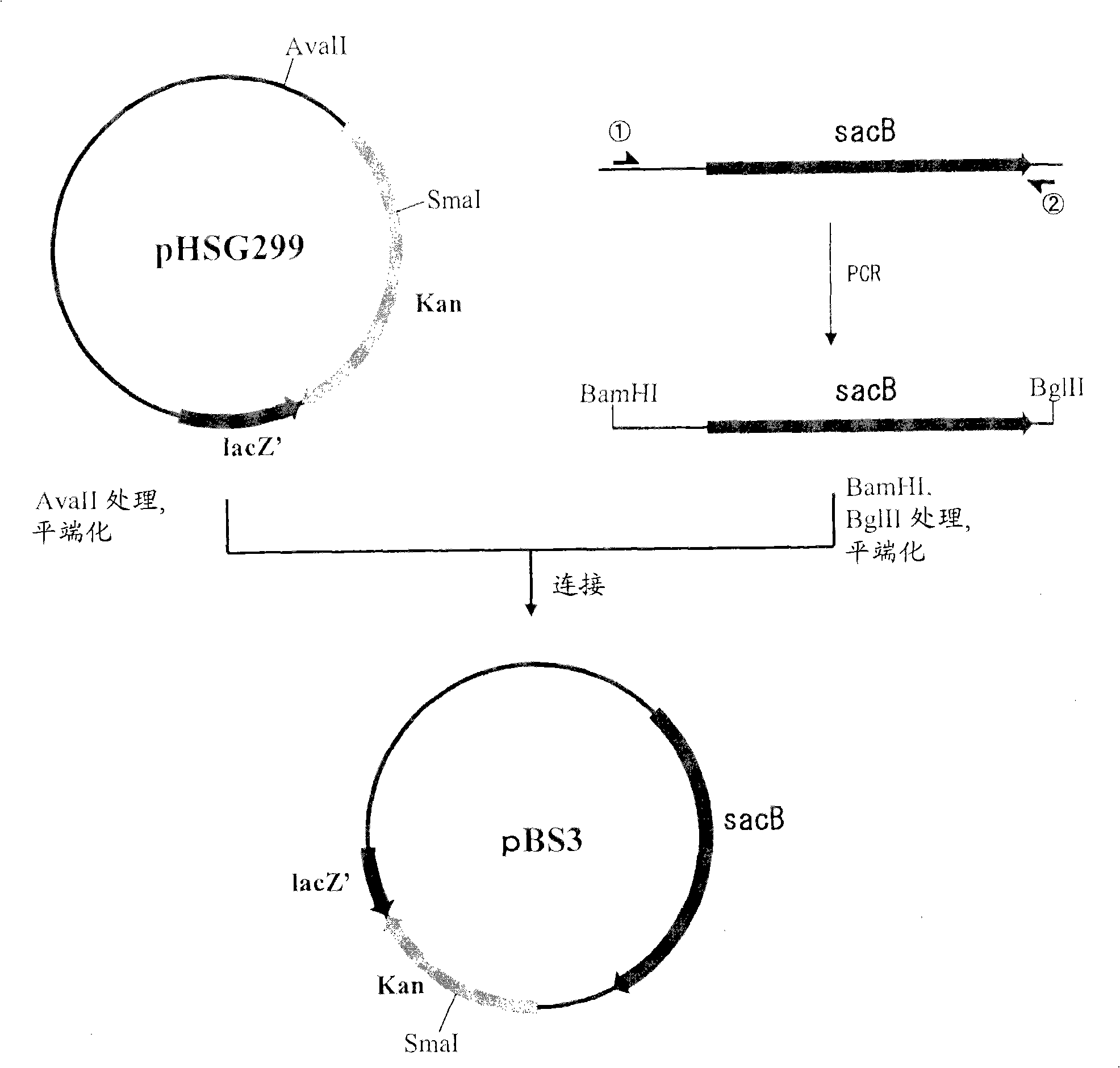
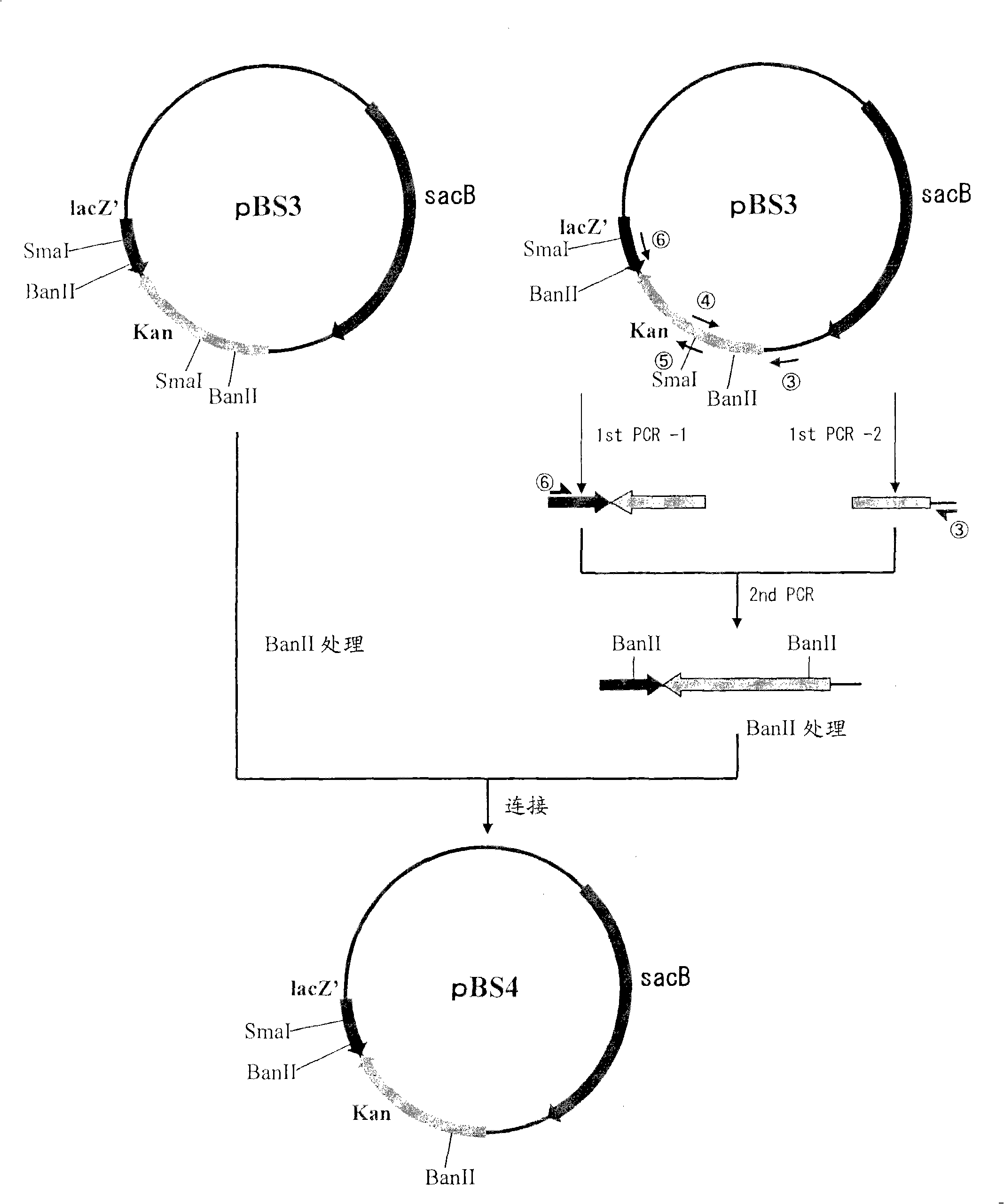
[0098] (A) Construction of pBS3
[0099] Plasmid pBS3 was obtained by PCR using chromosomal DNA of Bacillus subtilis as a template while using SEQ ID NOS: 1 and 2 as primers. The PCR reaction was carried out using LA taq (TaKaRa) as follows: one cycle of incubation at 94°C for 5 minutes, followed by a cycle of denaturation at 94°C for 30 seconds, annealing at 49°C for 30 seconds and extension at 72°C for 2 minutes was repeated 25 times. The PCR product thus obtained was purified by a conventional method, then digested with BglII and BamHI, and blunt-ended thereafter.
[0100] The fragment was inserted into the pHSG299 site digested with AvaII and blunt-ended. The DNA was used to transform competent cells of Escherichia coli JM 109 (TAKARA SHUZO CO., LTD.), and the transformed cells were spread to a culture medium containing 25 μg / ml kanamycin (hereinafter, abbreviated as Km). on LB medium, followed by overnight culture. Next, colonies that appeared were picked, a...
Embodiment 2
[0105]
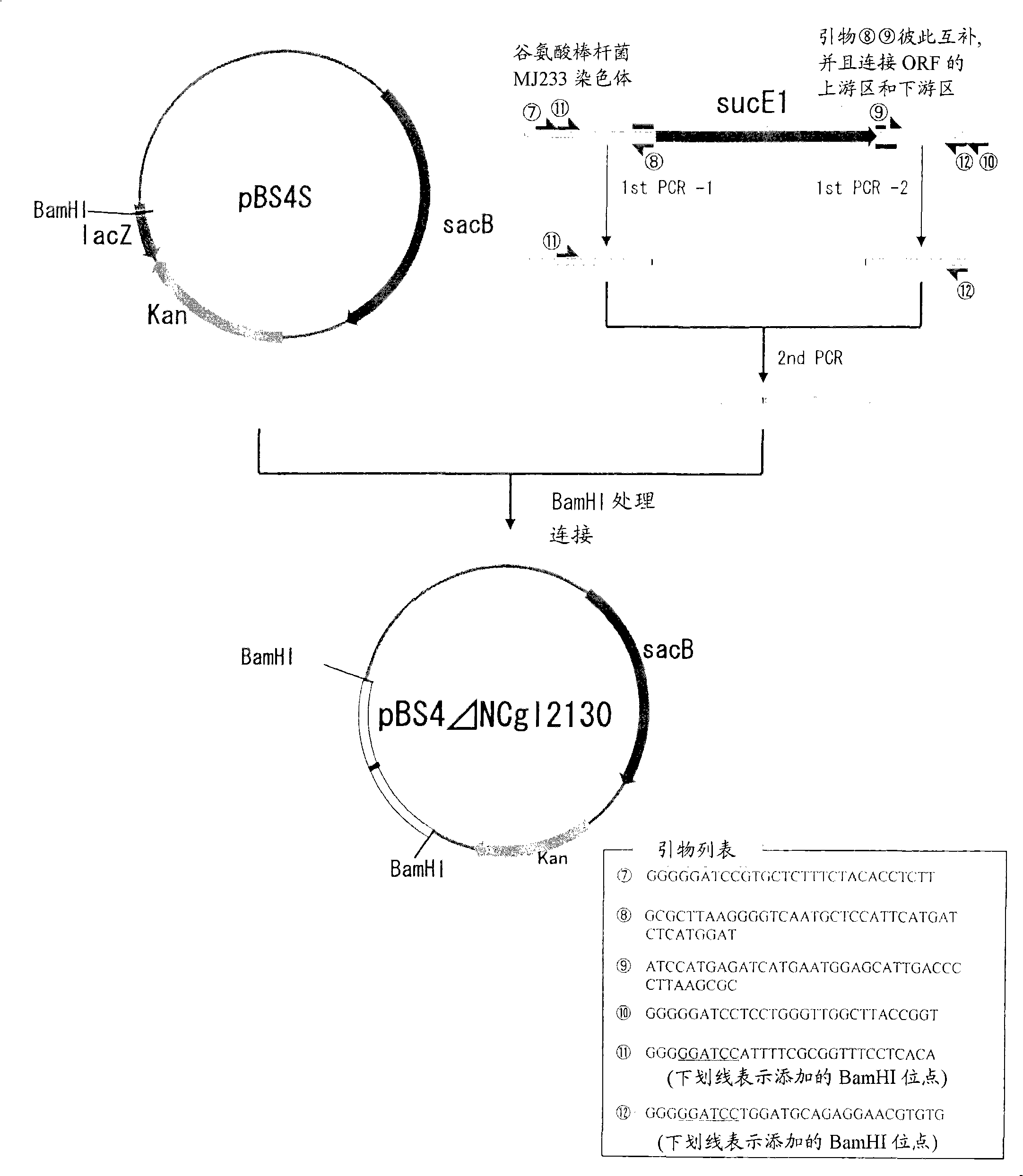
[0106] (A) Cloning of the fragment used to disrupt the sucE1 gene
[0107]Using synthetic DNA designed based on the nucleotide sequence around the NCgl2130 gene NCgl2130 of Corynebacterium glutamicum ATCC13032 (GenBank Database Accession No. NC#003450) that has been published as primers, a fragment of the gene in which the source is deleted is obtained by crossover PCR ORF from the sucE1 gene of Brevibacterium flavum MJ233 strain. Specifically, using the chromosomal DNA of the Brevibacterium flavum MJ233 strain as a template, and using the synthetic DNA of SEQ ID NOS: 7 and 8 in the Sequence Listing as primers, PCR was performed by a conventional method to obtain the amplification of the N-terminal region of the sucE1 gene. increase product. On the other hand, in order to obtain the amplified product of the C-terminal region of the sucE1 gene, using the genomic DNA of Brevibacterium flavum MJ233 as a template, and using the synthetic DNA of SEQ ID NOS: 9 and 10 in ...
Embodiment 3
[0112]
[0113] (A) Cloning of the sucE1 gene
[0114] A plasmid for amplifying the sucE1 gene derived from the Brevibacterium flavum MJ233 strain was constructed as follows. Use the synthetic DNA of SEQ ID NOS:13 and 14 in the sequence listing as primer, it is designed based on the nucleotide sequence near NCgl2130 of Corynebacterium glutamicum ATCC13032 (Genbank Database accession number NC003450), and uses the nucleotide sequence of Brevibacterium flavum MJ233 bacterial strain Genomic DNA was used as template for PCR. PCR was carried out using Pyrobest DNA Polymerase (TaKaRa) as follows: one cycle of incubation at 94°C for 3 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 2.5 minutes, resulting in PCR product of interest. The obtained PCR product was purified according to a conventional method and digested with Sse8387I, followed by insertion into the Sse8387I site of pVK9. The obtained ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com