Reagent kit for real time fluorescence quantitative PCR detection of bacillus pyocyaneus
A quantitative detection and Pseudomonas aeruginosa technology, applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve problems such as false positives, unsuitable for clinical diagnosis, PCR product contamination, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1: the development of Pseudomonas aeruginosa detection reagent
[0033] 1. Design of primers and probes: Through sequence comparison and analysis of all Pseudomonas aeruginosa nucleic acid sequences already available in the Genbank database and nucleic acid sequences reported in published literature at home and abroad, the main viruses related to Pseudomonas aeruginosa pathogenicity were analyzed. The force factor exotoxin A (ETA) gene was used as the amplification target site, and a highly conserved segment with no secondary structure was selected. According to the basic principles of primer and probe design, multiple pairs of primers and probes were designed by software and manually.
[0034] 2. Selection of clinical samples: According to relevant domestic and foreign literature reports, secretions or scrapings, sputum, urine, blood, cosmetics, and environmental monitors from clinical wounds, burns, corneal ulcers and other lesion areas can be selected. Whe...
Embodiment 2
[0045] Embodiment 2: Pseudomonas aeruginosa detection kit and its use
[0046] 1. Prepare a kit including the following components: 2 tubes of DNA extraction solution (500 μl / tube), 20 tubes of PCR amplification reaction solution (? μl / tube), 1 tube of negative quality control product (100 μl / tube), 1 tube of positive Control product (50μl / tube) 1 tube, quantitative standard (50μl / tube) 4 tubes.
[0047] 2. Specimen collection, transportation and storage
[0048] (1) Specimen collection: including various clinical specimens, such as blood, urine, sputum, pus, and puncture fluid, etc., as well as various specimens from the hospital environment, such as water, air, object surface sampling, etc., according to clinical Sampling is required for operation; it also includes cosmetic sampling, etc., and is operated in accordance with the national cosmetic microbial inspection standards. Sealed for inspection.
[0049] (2) Specimen storage and transportation: Specimens can be used f...
Embodiment 3
[0057] Example 3: Quantitative detection of clinical samples using Pseudomonas aeruginosa detection kit
[0058] The clinical samples come from the semi-solid culture of Nanjing Children's Hospital, and 50 μl of the upper layer of the culture is directly taken as the sample to be tested. Specimen processing, DNA extraction, PCR reaction and result analysis are carried out with reference to Example 2.
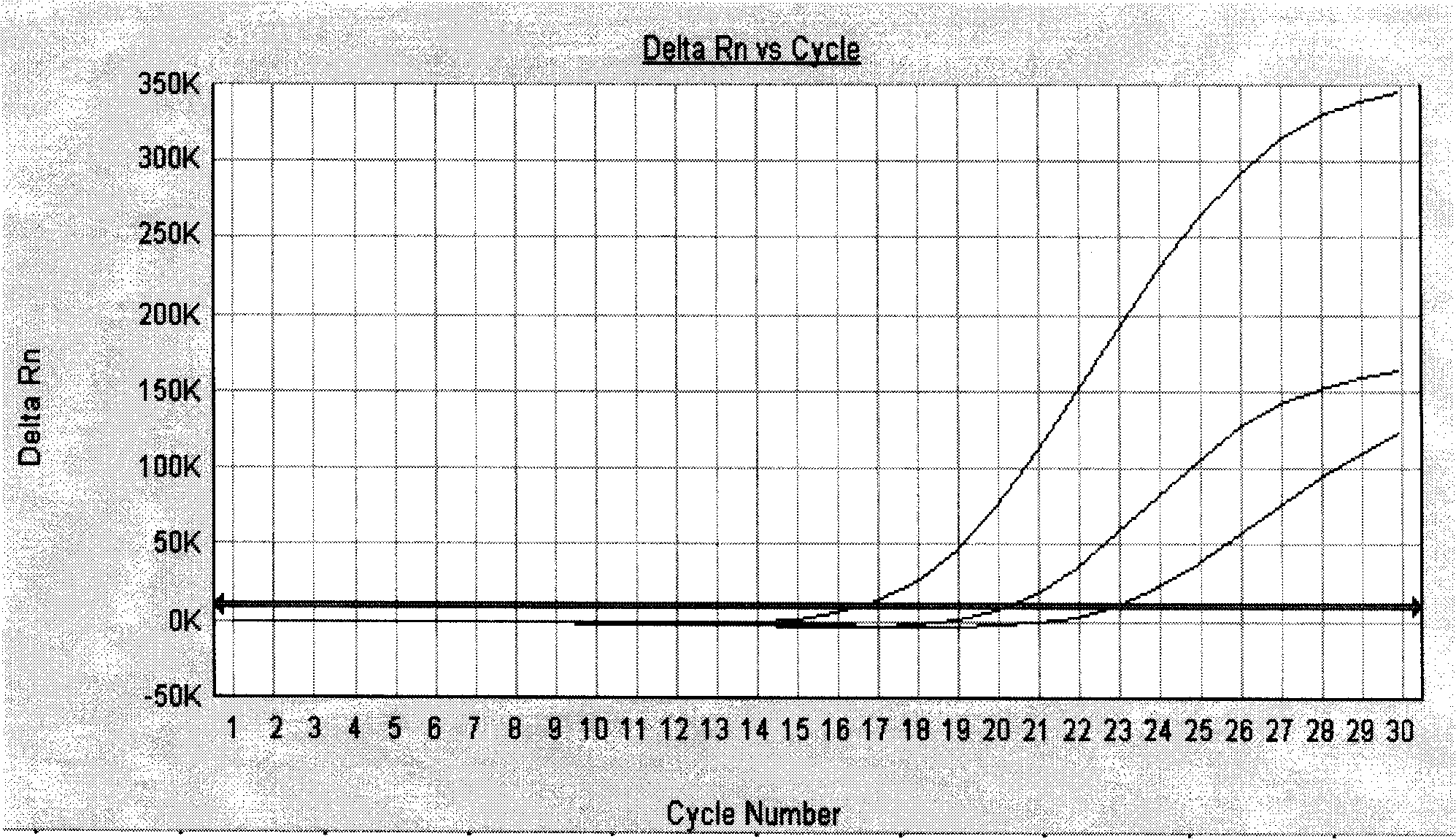
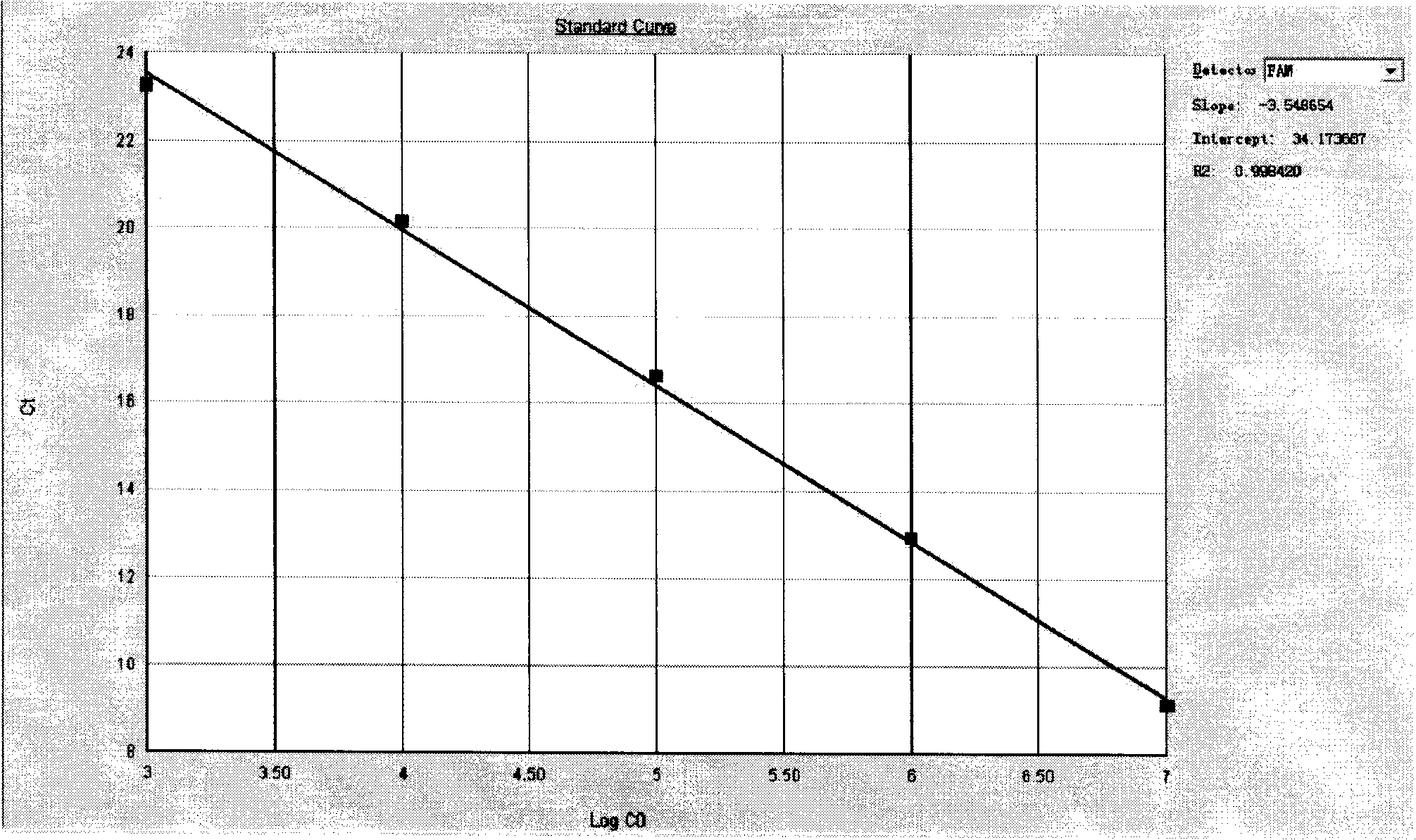
[0059] After the PCR reaction, adjust the analysis parameters according to the amplification curve to make the standard curve under the standard curve (Std curve) window reach the best (ie, the absolute value of the correlation value > 0.97), and then analyze the clinical samples. The test results of clinical samples are attached Figure 5 Shown: The Ct values of the amplification curves of the six clinically positive samples are 8.83, 9.63, 17.02, 19.01, 20.28, and 23.61, respectively. Combined with the obvious exponential growth period of the amplification curves, all of the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com