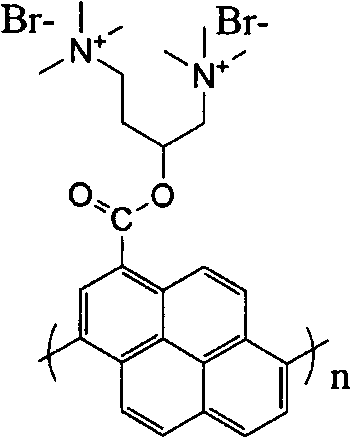
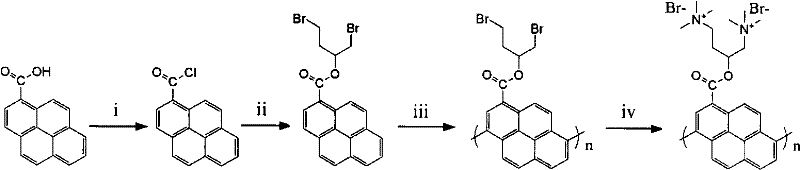
A fluorescence detection method that distinguishes single and dual -chain nucleotide
A new method, fluorescence analysis technology, applied in biochemical equipment and methods, fluorescence/phosphorescence, microbial determination/inspection, etc., can solve the problems of unfavorable π-electron interaction, small conjugated system, etc., to achieve low detection cost, Controllable effects of the preparation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Weigh an appropriate amount of the synthesized conjugated oligopyrene derivative and place it in a 15mL reagent bottle. Prepare a concentration of 10mmolL with ultrapure water -1 ; Tris-HCl buffer pH=7.4. Afterwards, add a certain volume of buffer solution into the reagent bottle containing the conjugated oligopyrene derivative to make the concentration 3.34×10 -6 mol L -1 , and pipette 3 parts of 5mL of the solution as the detection solution. Then, drop the ssDNA and dsDNA samples to be tested into the system. The final concentrations of ssDNA and dsDNA were both 8×10 -7 mol L -1 . The fluorescence response of the conjugated oligopyrene derivative detection solution to DNA is obtained by recording the fluorescence intensity of the fluorescence sensing system at 465 nm with a FL 4600 fluorescence instrument at 25 ° C, and the excitation wavelength is 340 nm. The experimental results are: ssDNA and dsDNA make the recovery values of the fluorescent signals of the...
Embodiment 2
[0026] Weigh an appropriate amount of the synthesized conjugated oligopyrene derivative and place it in a 15mL reagent bottle. Prepare a concentration of 10mmolL with ultrapure water -1 ; Tris-HCl buffer pH=7.4. Afterwards, add a certain volume of buffer solution into the reagent bottle containing the conjugated oligopyrene derivative to make the concentration 3.34×10 -6 mol L -1 , and pipette 3 parts of 5mL of the solution as the detection solution. Then, drop the ssDNA and dsDNA samples to be tested into the system. The final concentrations of ssDNA and dsDNA were both 3×10 -5 mol L -1 . The fluorescence response of the conjugated oligopyrene derivative detection solution to DNA is obtained by recording the fluorescence intensity of the fluorescence sensing system at 465 nm with a FL 4600 fluorescence instrument at 25 ° C, and the excitation wavelength is 340 nm. The experimental results are: ssDNA and dsDNA make the recovery values of the fluorescent signals of the...
Embodiment 3
[0028] Weigh an appropriate amount of the synthesized conjugated oligopyrene derivative and place it in a 15mL reagent bottle. Prepare a concentration of 10mmolL with ultrapure water -1 ; Tris-HCl buffer pH=7.4. Afterwards, add a certain volume of buffer solution into the reagent bottle containing the conjugated oligopyrene derivative to make the concentration 3.34×10 -6 mol L -1 , and pipette 3 parts of 5mL of the solution as the detection solution. Then, drop the ssDNA and dsDNA samples to be tested into the system. The final concentrations of ssDNA and dsDNA were both 8×10 -5 mol L -1 . The fluorescence response of the conjugated oligopyrene derivative detection solution to DNA is obtained by recording the fluorescence intensity of the fluorescence sensing system at 465nm with a FL 4600 fluorescence instrument at 25°C, and the excitation wavelength is 340nm. The experimental results are: ssDNA and dsDNA make the recovery values of the fluorescent signals of the con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com