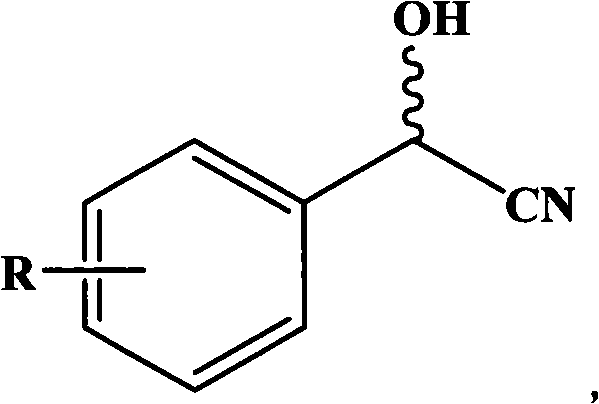
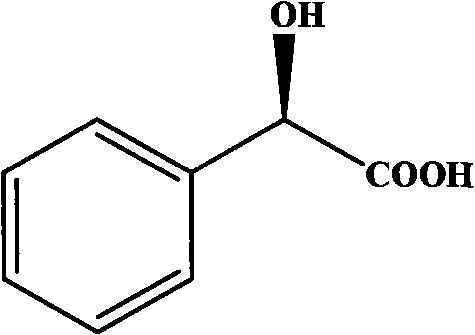
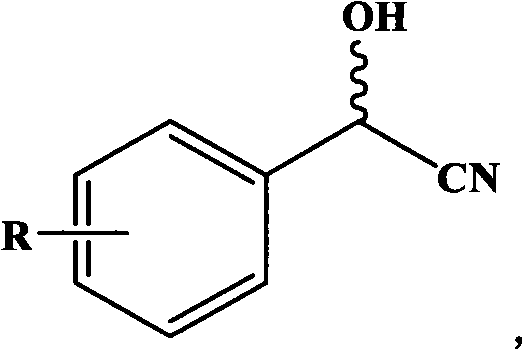
Gene for coding nitrilase of alcaligenes and method for preparing single enantiomorph of mandelic acid using same
A technology for nitrilase and alcaligenes, which is applied in the field of base sequences encoding nitrilase, can solve the problems of poor stability, low catalytic efficiency, low enzyme yield and the like, and achieves excellent effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Cultivation of Alcaligenes sp.ECU0401
[0034] Alcaligenes sp.ECU0401 was cultured in a liquid medium with the following composition at 30° C. and 180 rpm for 48 hours with shaking to prepare cells expressing nitrilase activity.
[0035] Culture Medium Composition
[0036] Glycerin: 10g / L; peptone: 15g / L; yeast extract: 8g / L; KH 2 PO 4 : 4g / L; NaCl: 2g / L; MgSO 4 : 0.2g / L; FeSO 4 : 0.05g / L; pH 7.0.
[0037] The fermentation broth was centrifuged at 12,000×g for 10 min to recover the cells, washed twice with saline, and stored in a refrigerator at 4°C for future use.
Embodiment 2
[0038] Example 2 Preparation of Alcaligenes Genomic DNA
[0039] After activation, the bacteria cells Alcaligenes sp.ECU0401 were inoculated in 1.5ml fermentation medium, cultured at 30°C for 12 hours, and the cells were collected by centrifugation (12000rpm, 3min). The cells were washed twice with TE solution (10000 rpm, centrifuged for 3 min), and resuspended in 300 μl TE solution. Add 200 μl of 10% SDS and 5 μl of 20 mg / ml proteinase K, mix well, and incubate at 30° C. for 2 hours. After the bacterial cells burst, a large amount of polysaccharides are released, add a mixture of 500μl 5mol / L sodium chloride and 500μl CTAB / NaCl solution, invert the centrifuge tube up and down, mix well, incubate at 55°C for 10min, then take it out and mix again , and incubated at 55°C for another 10min. Cool the mixture in an ice bath, add an equal volume of phenol chloroform (phenol: chloroform: isoamyl alcohol = 25: 24: 1) to extract twice, centrifuge at 12000 rpm for 10 min. Take the su...
Embodiment 3
[0040] Example 3 Preparation of Genomic DNA Library of Alcaligenes sp.ECU0401
[0041] The genomic DNA obtained in Example 2 was completely digested with the restriction endonuclease Sau I, separated by agarose gel electrophoresis, segmented according to the length of the DNA, and the DNA fragments above 2 kb were collected, and the 5' end was restricted. The endonuclease Sau I digested and dephosphorylated vector pUC18 was ligated, and the resulting plasmid library was transformed into Escherichia coli DH5α, and the transformants were spread on LB agar plates containing ampicillin (100 μg / ml) and kept at 37°C. cultured to form single colonies.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com