Preparation method of maltooligosyltrehalose hydrolase gene sequence and recombinant protein thereof
A technology based on trehalose hydrolase and maltooligosaccharides, which is applied in the biological field and can solve problems such as limited application and unsatisfactory results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
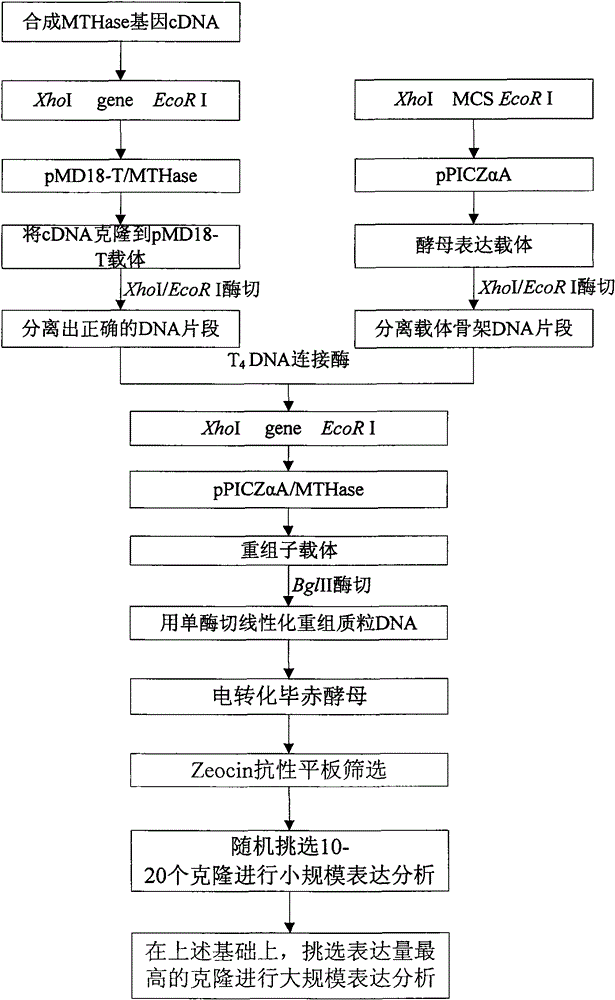
[0043] Example 1 Artificial modification and synthesis of malto-oligosaccharyl trehalose hydrolase (MTHase) gene
[0044] According to the above coded amino acid sequence, the forward and reverse strand DNA primers (as shown in SEQ ID NO: 6 to SEQ ID NO: 47) for gene splicing are designed according to the principle of yeast codon preference. XhoI restriction site is added to the 5'end, and EcoR I restriction site sequence is added to the 3'end to facilitate connection with the vector.
[0045] Prepare primers with ultrapure water to make the concentration of primers: 100pmol / μL. Take 1μL of the primers respectively. After mixing, let them react in boiling water at 100℃ for 15 minutes, then immediately bath them on ice; add 5μL of 10X T4DNA ligase Buffer, T4 Polynucleotide Kinase 5μL, and add water to the total The volume is 45μL, 37°C for 30 minutes; 65°C for 20 minutes, ice bath for 2 minutes; 5μL T4 DNALigase is added, 16°C for 2 hours, 65°C for 20 minutes.
[0046] Take 1μL of th...
Embodiment 2
[0047] Example 2 Construction of recombinant expression plasmid and screening of positive clones
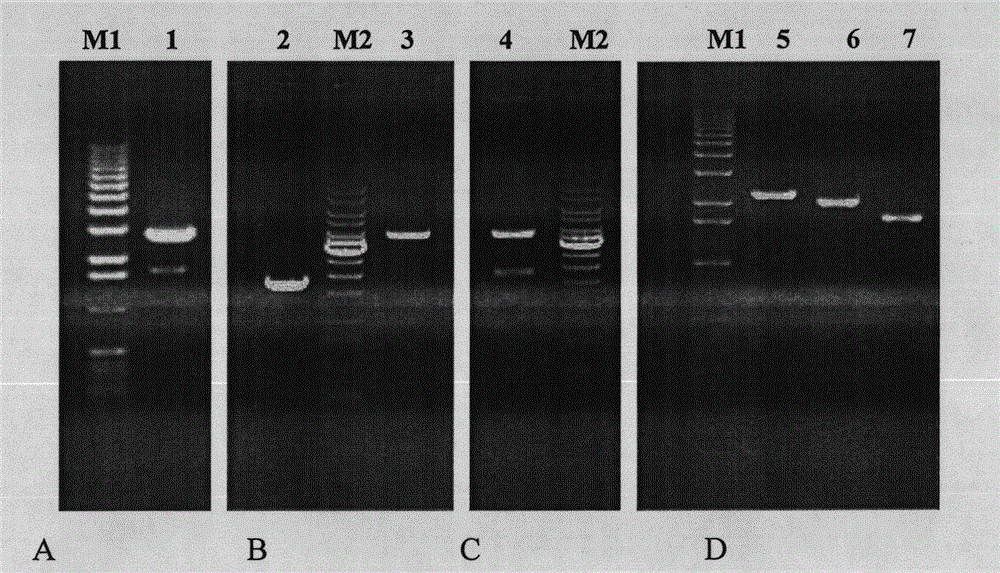
[0048] The PCR product finally obtained in Example 1 was purified and recovered, and connected to pMD18-T vector (Takara, Dalian Bao Bioengineering Company). After sequence analysis, the correct positive clone pMD8-T / MTHase was selected, and the sequence obtained was SEQID NO : 4. Double-enzyme digestion with XhoI / EcoRI, isolate and recover the MTHase gene fragment of about 1.7kb, connect it to the pPICZαA yeast expression vector (Invitrogen, USA) digested with XhoI / EcoRI, and screen positive clones pPICZαA / MTHase. After the positive cloning vector was digested with BglII single enzyme, the Pichia pastoris SMD1168H yeast was transformed, and the positive clone strain was selected on the high concentration Zeocin-YPD solid medium ( Pichia pastoris SMD1168H / pPICZαA-MTHase ), and perform further PCR identification and confirmation.
Embodiment 3
[0049] Example 3 Medium and culture expression conditions
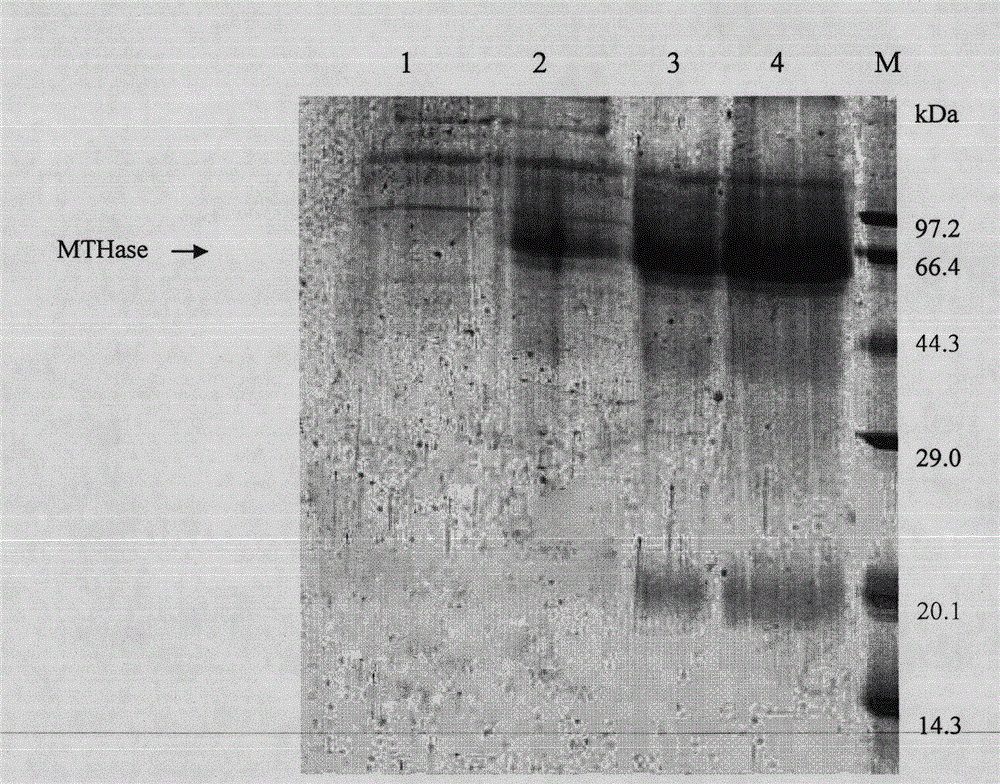
[0050] The positive strain described in Example 2 ( Pichia pastoris MD1168H / pPICZαA-MTHase ) Inoculated in YPD medium, cultivated at 30°C for 12-24 hours, and the shaker speed is 240 rpm; when the concentration of the fermentation broth reaches OD 600 =5.0, transfer the inoculum to BMG medium at a volume ratio of 1:100, temperature 30℃, shaker speed 240rpm, culture for 28-35h; centrifuge at 5000g for 5min, collect the bacteria, and wash the bacteria with PBS 1-2 times, transfer to BMM liquid medium, when the concentration of the fermentation broth reaches OD=0.5-1.0, induce culture, the induction temperature is 25℃, the shaker speed is 180rpm, and 5- is added to the medium every 12h. 10mL / L of 0.5% methanol, incubate for 72 hours;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com