Expression vector for stably indicating cell autophagy activities, establishing method and application method thereof
A technology for expressing vectors and cells, which is applied in the field of biomedicine, can solve problems such as the distortion of fluorescent spot observation, difficulty in screening stable strains and amplification, increase the difference in biological characteristics between transfected cells and mother cells, and achieve quantitative analysis of autophagy The method is simple and easy to use, the indication of autophagy is sensitive and reliable, and the effects on a wide range of cell types
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: This example is used to illustrate the construction process of an expression vector stably indicating autophagy activity of cells, and the application of expression vectors to establish autophagy indicator cells and quantitative analysis applications;
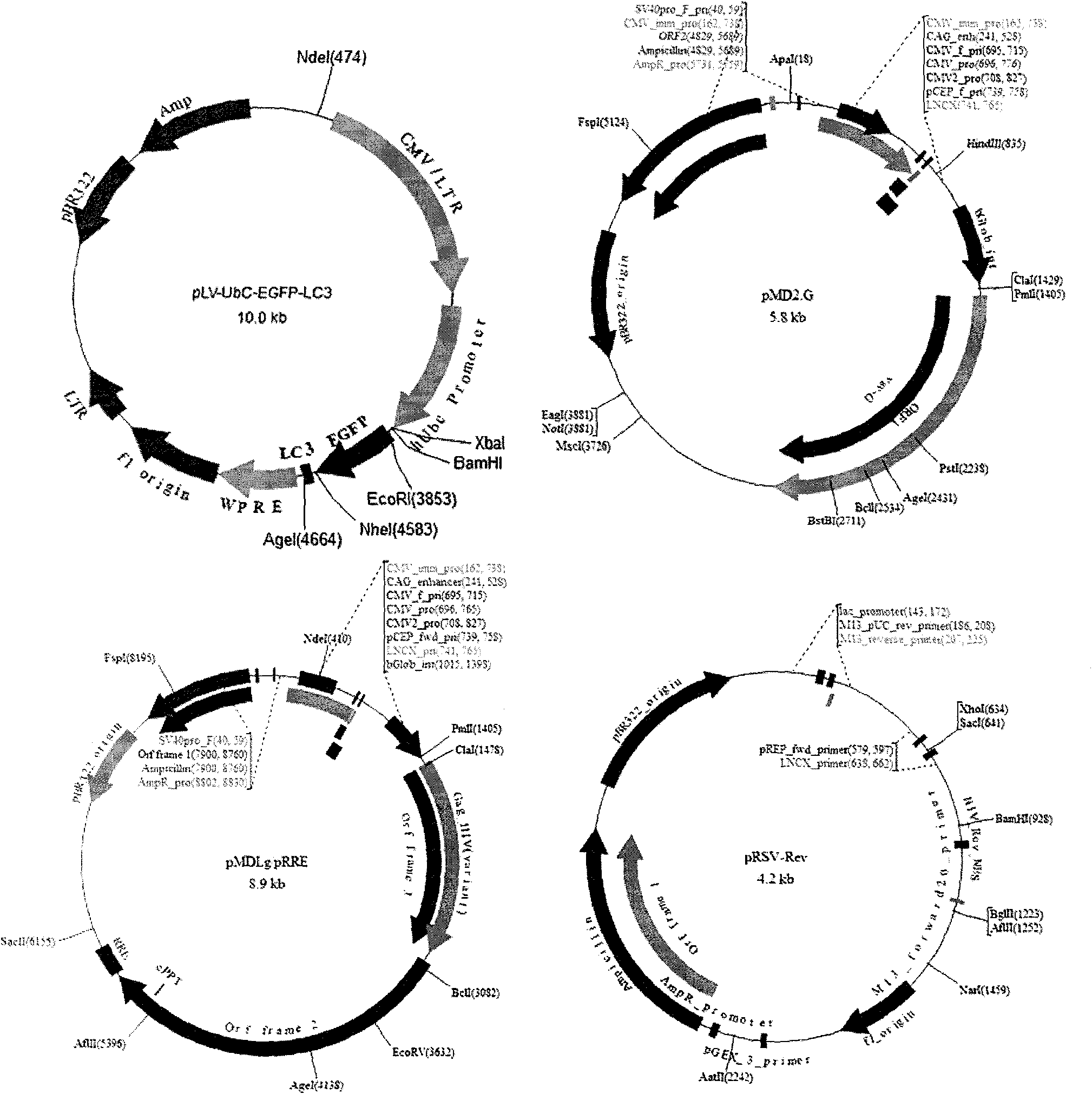
[0022] (1) Expression vector construction: ① Expression plasmid construction: NCBI obtained LC3 gene sequence information and artificially synthesized LC3 gene. EcoR V digested the pUC57 plasmid and synthesized the LC3 gene, and inserted the LC3 gene fragment into the pUC57 vector to construct pUC57-LC3. Transform competent cells, screen clones, extract plasmids by electrophoresis and KpnI, HindIII double enzyme digestion identification, and sequence to confirm the insertion product. The clones with correct sequencing were re-inoculated and amplified for plasmid extraction. pUC57-LC3 was double digested with Nhe I and Age I, and the 384bp LC3 gene fragment was recovered by gel after electrophoresis. The pLV-...
Embodiment 2
[0025] Embodiment 2 to 5 are in order to further illustrate the concrete application of the present invention
[0026] Example 2
[0027] Apply the expression vector of the present invention to infect difficult-to-transfect cells to establish corresponding autophagy indicator cell lines
[0028] Some cell lines are difficult to transfect by common transfection methods (such as liposome method), and the transfection rate is usually less than 10%. If the cell line is sensitive to the toxicity of the transfection reagent, the transfection is successful. It is difficult to carry out follow-up experiments. At the same time, the background autophagy of the traditional transfection method is high ( Figure 4- left). In this example, HCCLM3 liver cancer cells, Caco-2 colon cancer cells and NIH3T3 fibroblasts are represented by three difficult-to-transfect cells, and the expression vectors of the present invention are used to establish corresponding autophagy indicator cells (while ...
Embodiment 3
[0030] Application of the present invention to carry out research on the correlation between drug resistance of liver cancer cells and autophagy
[0031] Tumor resistance to chemotherapeutic drugs is an important obstacle in tumor treatment, and its related research has always been a hot spot. Studies in recent years have shown that autophagy is an important mechanism of tumor cell chemotherapy resistance, and inhibition of autophagy increases chemotherapy sensitivity. The in vitro study of tumor drug resistance needs to detect the changes of autophagy of different cell lines under the action of different chemotherapeutic drugs and different drug doses. The workload of detection by conventional Western blot, transmission electron microscopy and other methods is very large, and the application of the present invention The workload can be greatly reduced and rapid screening can be completed. In this case, the effects of 8 different chemotherapeutic drugs at standard blood level...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com