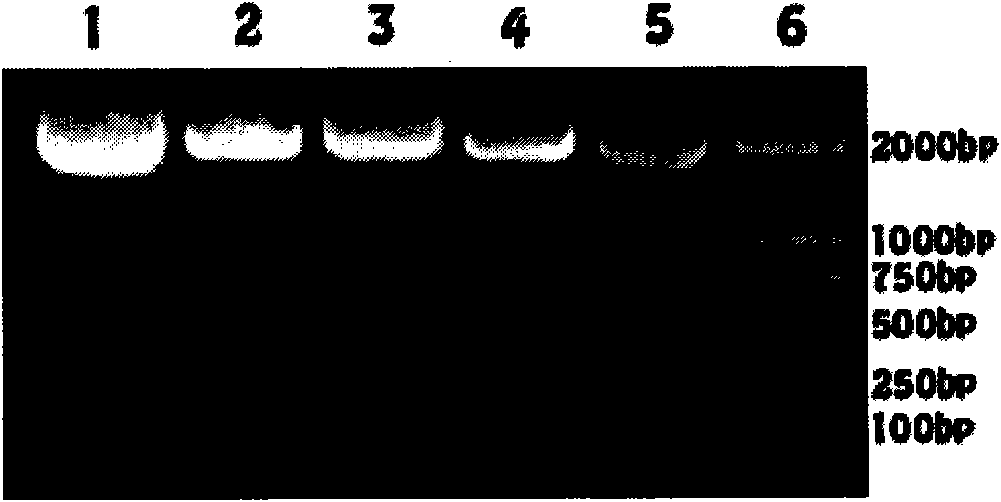Recombinant pseudomonas putida, and construction method and application thereof
A technology of Pseudomonas putida and a construction method, applied in the field of recombinant Pseudomonas putida and its construction, and can solve the problems of unrealistic phosphorus removal and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Acquisition of ppk gene.
[0034] The total DNA of E.coli DH5α was extracted. For specific methods, refer to "Environmental Microbial Experimental Technology" (Beijing: China Environmental Science Press, 2004.71-72). Using the E.coli DH5a genome as a template, primers were designed according to the DNA sequence of the ppk gene (Genebank accession number: L03719), and the ppk gene was amplified by PCR for transgenic operation.
[0035] Forward primer: 5′-ATG CAG ATG GGT CAG GAA AAG CTA TAC AT-3′;
[0036] Reverse primer: 5'-GGT ACC CAG GTT GTT CGA GTG ATT TGA-3'.
[0037] The PCR reaction conditions were: pre-denaturation at 95°C for 5 min; denaturation at 94°C for 30 s, annealing at 60°C for 45 s, extension at 72°C for 2 min, and the number of cycles was 30.
[0038] The enzyme used in PCR was PrimeSTAR purchased from TAKARA (JAPAN) TM HS DNA polymerase. A fragment of about 2 kb was amplified. The enzymes used in PCR are high-fidelity enzymes and the prod...
Embodiment 2
[0039] Example 2: Construction of vectors.
[0040] The PCR product obtained in Example 1 was subjected to a phosphorylation reaction, and phosphorus was added to the 5' end. Plasmid pBBR1MCS-2 [Kovach, M., P. Elzer, et al. (1995). "Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes." Gene 166(1):175-176.] After EcoR V digestion, dephosphorylation reaction was performed to avoid self-ligation. The PCR product (ppk) treated above was reacted overnight with plasmid pBR1MCS-2. Single clones were picked for sequencing. The recombinant plasmid pBR1MCS-2-ppk inserted in the forward direction was obtained. Transformed into the host E.coli DH5α by electroporation for replication. The recombinant plasmid pBBR1MCS-2-ppk was extracted and used as a template to design primers containing Not I restriction sites.
[0041] Forward primer: 5′ GCG GCC GC C GTT GCG TCG CGG TGC A-3' (Not I);
[0042] Reverse primer: ...
Embodiment 3
[0046] Example 3: Triparental conjugation.
[0047] The recombinant plasmid pUTmini-Tn5-ppk was transformed into the donor strain E.coli SM10(λpir) by electric shock (for the preparation of electric shock transformation and competent cells, please refer to the "Molecular Cloning Guide" [3 rd , Beijing: Science Press. 2002.]).
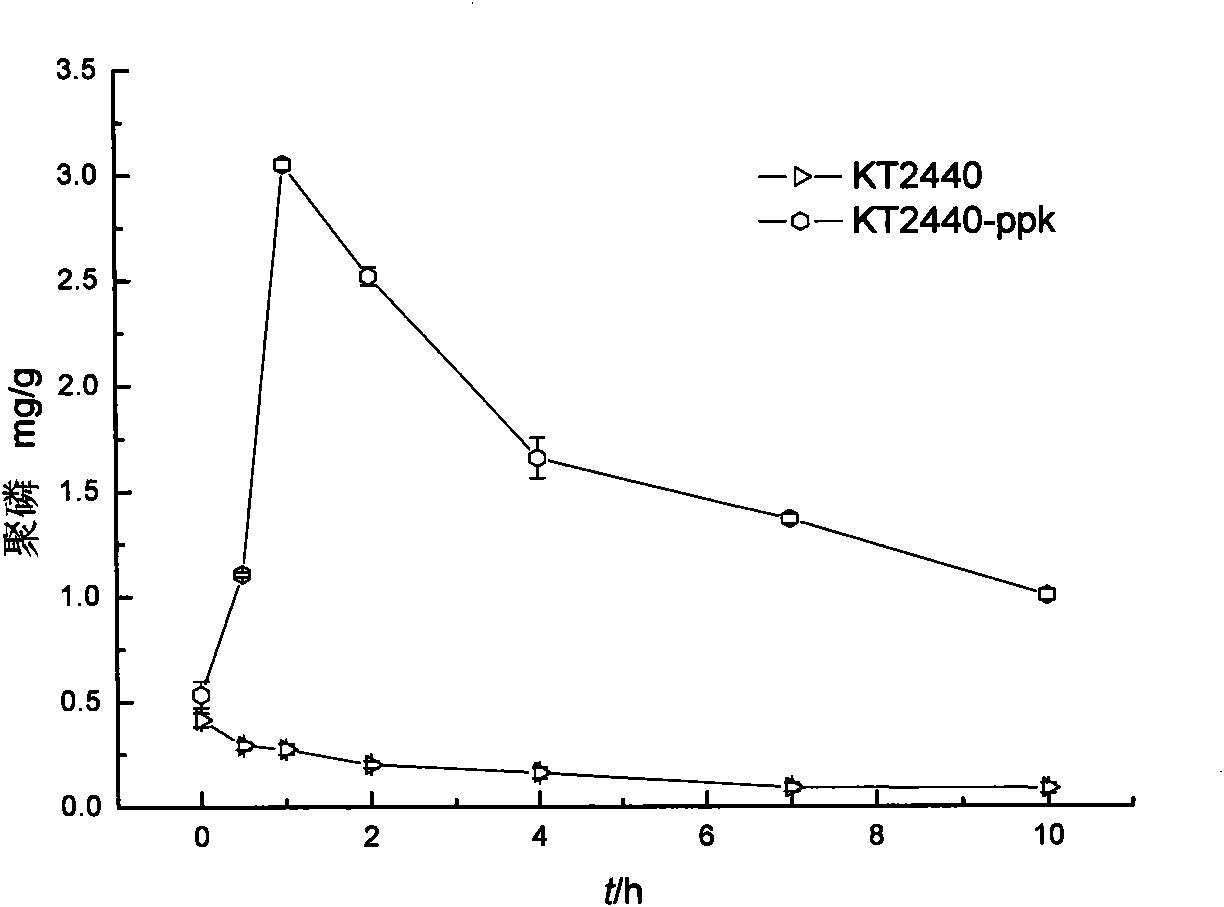
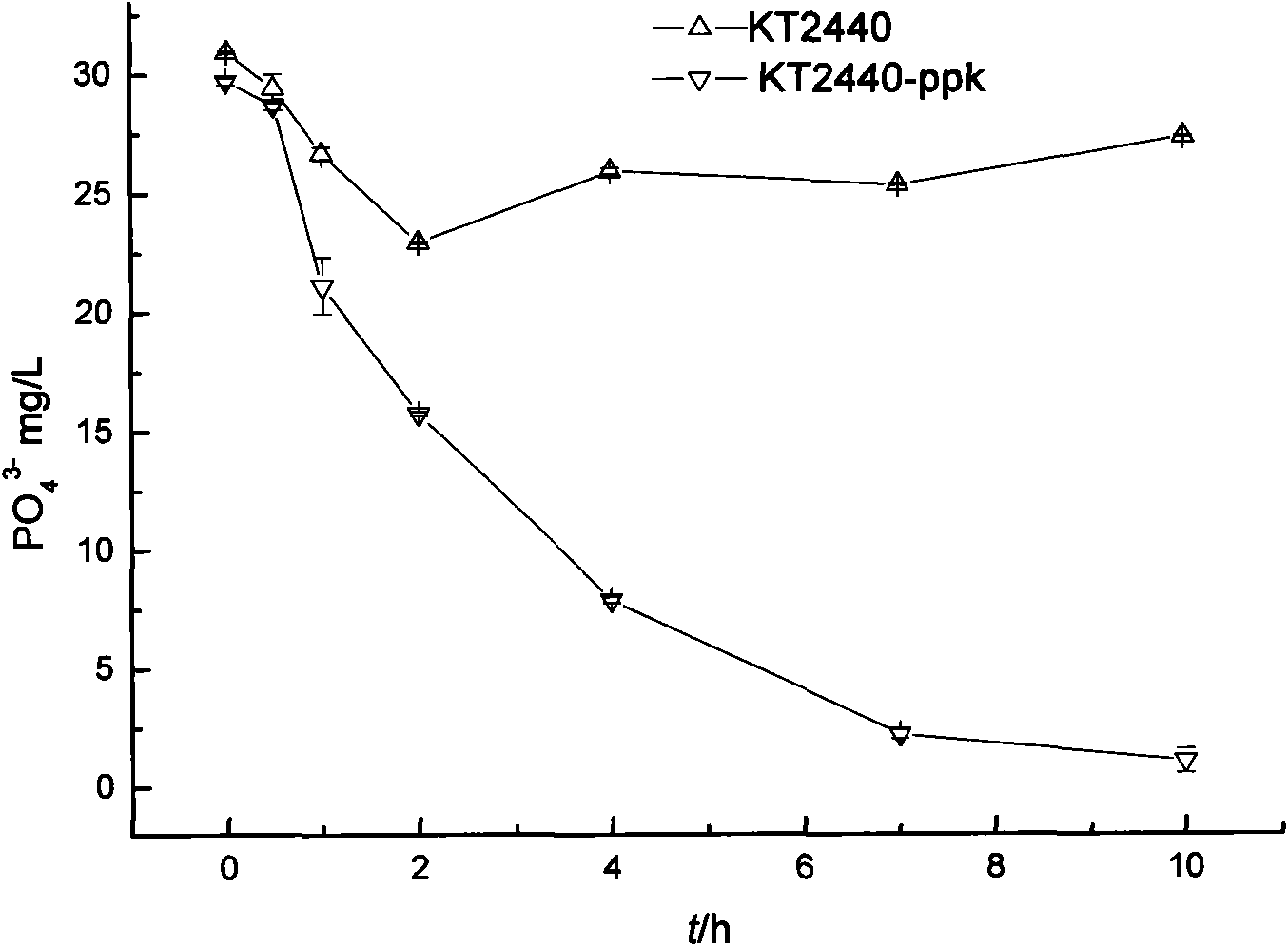
[0048] The recipient bacterium (Pseudomonas putida KT2440), the donor bacterium [E.coli SM10(λpir) / pUTmini-Tn5-ppk] containing the corresponding plasmid and the auxiliary bacterium E.coli DH5α(pRK600) were cultured overnight in LB medium. And add chloramphenicol (final concentration is 25 μg / mL) in KT2440 medium, add the final concentration of 10 μg / mL kanamycin and 25 μg / mL chloramphenicol in the culture medium of donor bacteria and auxiliary bacteria . The recipient bacteria were incubated at 42°C for 15 minutes to inactivate the restriction system. Then add 2 mL of donor bacteria and 2 mL of auxiliary bacteria to 1 mL of recipient bacteria and mix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com