One-stop PCR (Polymerase Chain Reaction) method
A station type, dye mixture technology, applied in the field of molecular biology, can solve problems such as harm to the health of experimenters, low PCR amplification efficiency, consumption of large time and energy, etc., to achieve simple and easy preparation, improve efficiency and sensitivity. , the effect of reducing operation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1, the preparation (100ml) of 5 * compatible dye mixture
[0046] (1) Add 80ml of PCR grade pure water to a clean beaker;
[0047] (2) Weigh 10g of Ficoll 400 dry powder, add it into the above pure water, pay attention to mixing with a magnetic stirrer while adding, do not make Ficoll 400 agglomerate, otherwise it will affect the sedimentation effect of subsequent spotting;
[0048] (3) Take by weighing 0.02g cresol red, join in the solution of step (2), magnetic stirring aids dissolution;
[0049] (4) Take by weighing 0.08g tartrazine, join in the solution of step (3), magnetic stirring aids dissolution;
[0050] (5) Put the solution in step (4) in a 70°C incubator and heat it at a high temperature to help dissolve it for 1 hour. During this period, shake vigorously every 10 minutes or so. At this time, you can see that the solution is transparent yellow-red, which is normal color;
[0051] (6) After the solution in step (5) is taken out and cooled, add a...
Embodiment 2
[0057] Example 2, 2×PCR master mixture preparation
[0058] The 2×PCR master mixture was prepared, and the ingredients and dosage are shown in the table below:
[0059] 5× compatible dye mix 4ul
[0060] 10×reaction buffer 2ul
[0061] dNTP (10mmol / l each) 0.5ul
[0062] Taq DNA polymerase (5units / ul) 0.2ul
[0063] PCR grade pure water 3.3ul
[0064] Total volume 10ul
[0065] Mix the components sequentially according to the order in the above table, and the preparation can be scaled up in proportion.
[0066] Among them, Taq DNA polymerase, dNTP, and 10× reaction buffer were purchased from Shanghai Shenergy Biotechnology Co., Ltd.
[0067] The 2×PCR main mixture can be divided into 1.5ml centrifuge tubes, 1ml / tube, and can be stored at -20°C for a long time under dark conditions. The normal color of the 2X PCR master mix is dark red.
Embodiment 3
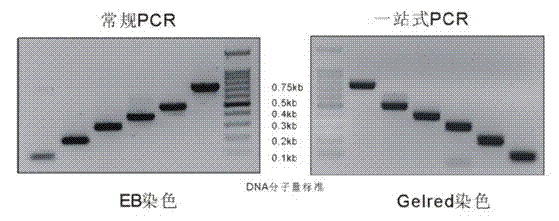
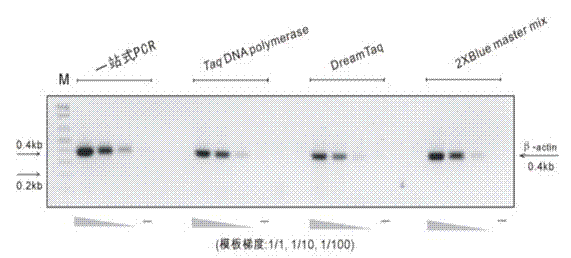
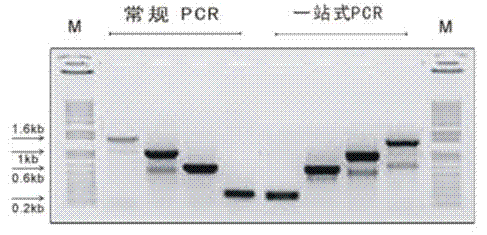
[0068] Example 3 One-stop PCR does not affect the electrophoretic mobility of the product ( figure 1 )
[0069] (1) Using human Hela cell cDNA as a template (prepared with 2 μg total RNA), amplify 0.1kb, 0.2kb, 0.3kb, 0.4kb, 0.5kb, 0.75kb fragments of β-actin cDNA. The preparation of the reaction system is as follows:
[0070] 2×PCR Master Mix 10ul
[0071] Upstream primer (10uM) 0.5ul
[0072] Downstream primer (10uM) 0.5ul
[0073] Template cDNA 1μl
[0074] Water 8ul
[0075] Total volume 20ul
[0076] (2) The reaction conditions are:
[0077] Pre-denaturation at 94°C for 2 minutes
[0078]
[0079] 72℃ 5min total extension
[0080] (3) After the PCR reaction, 5 μl of the product was taken and detected by electrophoresis on 2% agarose gel.
[0081] Depend on figure 1 The results showed that one-stop PCR had no effect on the electrophoretic mobility of PCR products.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com