Gene chip for detecting mutation of 18 loci of susceptibility genes of type 2 diabetes
A type 2 diabetes and gene chip technology is applied in the field of gene chips for rapid detection of multiple site mutations in type 2 diabetes susceptibility genes, which can solve the problems of limited detection gene sites, cumbersome operations, and high cost, and achieve consistent fluorescence intensity. The effect of good sex and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Probe and Primer Design
[0051] The present invention screens type 2 diabetes susceptibility gene site mutations, designs related probes, and obtains a group of probes with high hybridization specificity and high accuracy after repeated tests, screening and verification. The probe sequences are shown in Table 1. Show.
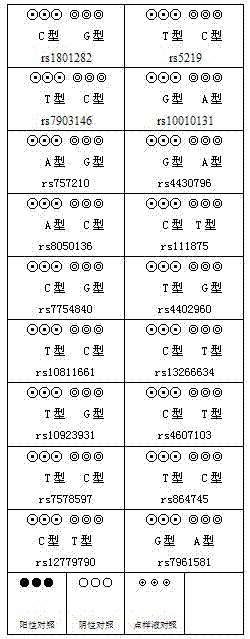
[0052] Table 1 The wild-type and mutant probe sequences of 18 sites
[0053]
[0054]
[0055] When the probe is synthesized, 15 more poly-Ts are synthesized at the 5' end, and are modified by amino groups (using standard phosphoramidite chemical method, 5' or 3' amino modification with 5'-Amino-Modifier C6, TFA- protected is introduced in the last step after synthesis). The poly-T modified probe required for the chip (the probe was synthesized by InvitrogenG) was resuspended with TE Buffer (100mM Tris-Cl pH 8.0, 10mM EDTA pH 8.0) to a solution with a concentration of 50μM, and the concentration was 1:1 before sample application. The ratio was ...
Embodiment 2
[0061] Glass medium gene chip preparation
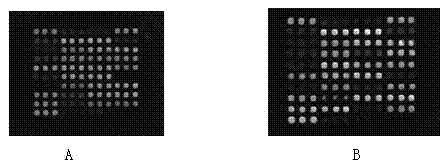
[0062] Aldehylation modification of the glass slide; resuspend the pre-synthesized probe (the probe was synthesized by InvitrogenG) with TEBuffer (100mM Tris-Cl pH 8.0, 10mM EDTA pH 8.0) to a solution with a concentration of 50uM, press 1 : 1 ratio mixed with spotting buffer (Micro Spotting Solution Plus 2X, Telechem), the final concentration is 25 μ M; figure 1 ) are arranged in a 96-well plate; PBS solution washes the spotting needle of the spotting instrument (SpotBot3, Telechem), calibrates, and presses figure 1 Matrix spotting; room temperature hydration fixation.
Embodiment 3
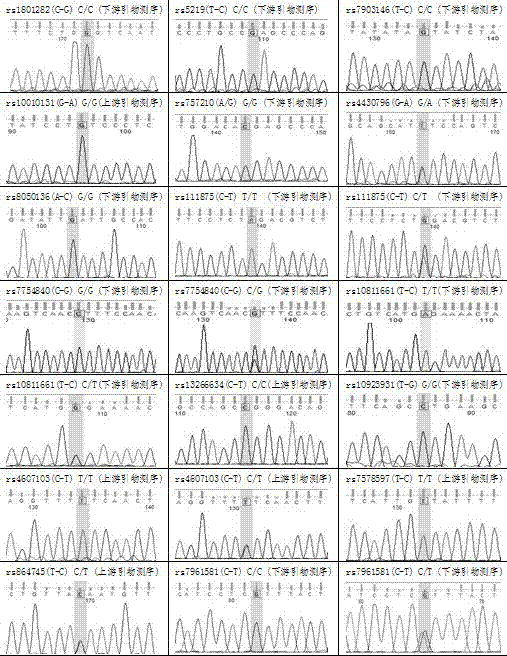
[0064] Detection methods of susceptibility loci
[0065] (1) Sample DNA extraction
[0066] Peripheral blood DNA was obtained, and genomic DNA was extracted using a blood genome DNA extraction kit (blood genome extraction kit DP318-02, TIANGEN) according to the manufacturer's instructions.
[0067] (2). Target DNA amplification, hybridization, detection:
[0068] The PCR reaction solution is composed of 10×buffer, 10μmol / L forward primer, 10μmol / L reverse primer, 25mmol / LMgCl 2 , 10mmol / L dNTPs (containing 0.5nM Cy3-dCTP, provided by Amersham Company) and sterile double distilled water. Amplification conditions are pre-denaturation at 95°C for 3 min, 35 cycles (denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s), extension at 72°C for 7 min; denaturation at 100°C, quenching; pre-hybridization at 42°C for 30 min to block non-specificity Binding; preheated denatured Cy3-DNA and hybridization solution (DIG Easy Hyb, Roche) were mixed at a rat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com