Kit and method for rapidly detecting DNA methylation
A methylation and kit technology, applied in the field of biomedical detection, can solve the problems of incomplete modification, low experiment repeatability, and pain for the person to be tested, so as to reduce operation steps, avoid cross-contamination, and save time-consuming consumables. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] The preparation of embodiment 1 kit
[0067] The three solutions were prepared according to conventional methods.
[0068] Recipe 1 is as follows:
[0069] (1) Modification solution: 5M sodium bisulfite, 5M ammonium bisulfite, 5mM hydroquinone, 2mM MBT (2-mercaptobenzothiazole), 4M guanidine isothiocyanate, 50mM Tris-HCl pH 5.5;
[0070] (2) Cleaning solution: 35% ethanol, 400mM sodium hydroxide, 200mM sodium chloride;
[0071] (3) Washing solution: 10 mM Tris pH 6.8, 80% ethanol.
[0072] The solvent of the three solutions is water.
[0073] Assemble the kit after aliquoting the modification solution, cleaning solution, and washing solution.
[0074] Recipe 2 is as follows:
[0075] (1) Modification solution: 4M sodium bisulfite, 3M ammonium bisulfite, 7mM hydroquinone, 5mM MBT (2-mercaptobenzothiazole), 2.6M guanidine isothiocyanate, 40mM Tris-HCl pH 6.0;
[0076] (2) Cleaning solution: 30% ethanol, 500mM sodium hydroxide, 200mM sodium chloride;
[0077] (3) Wa...
Embodiment 2
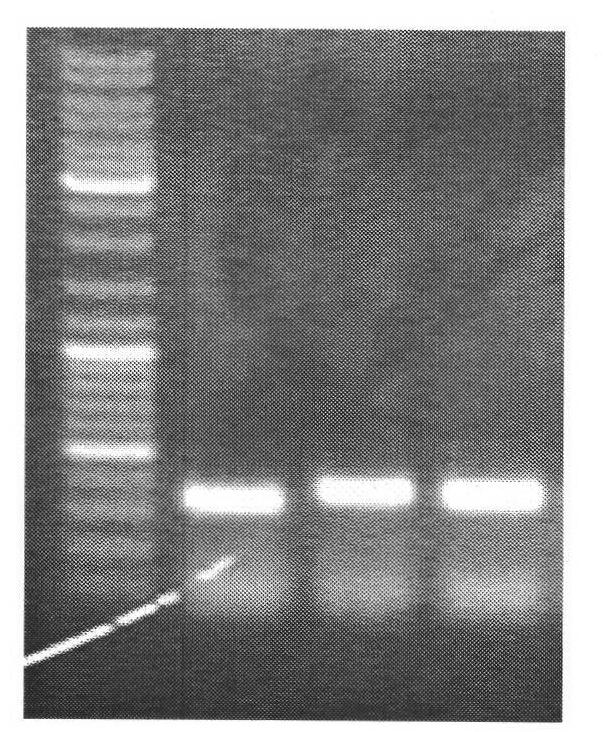
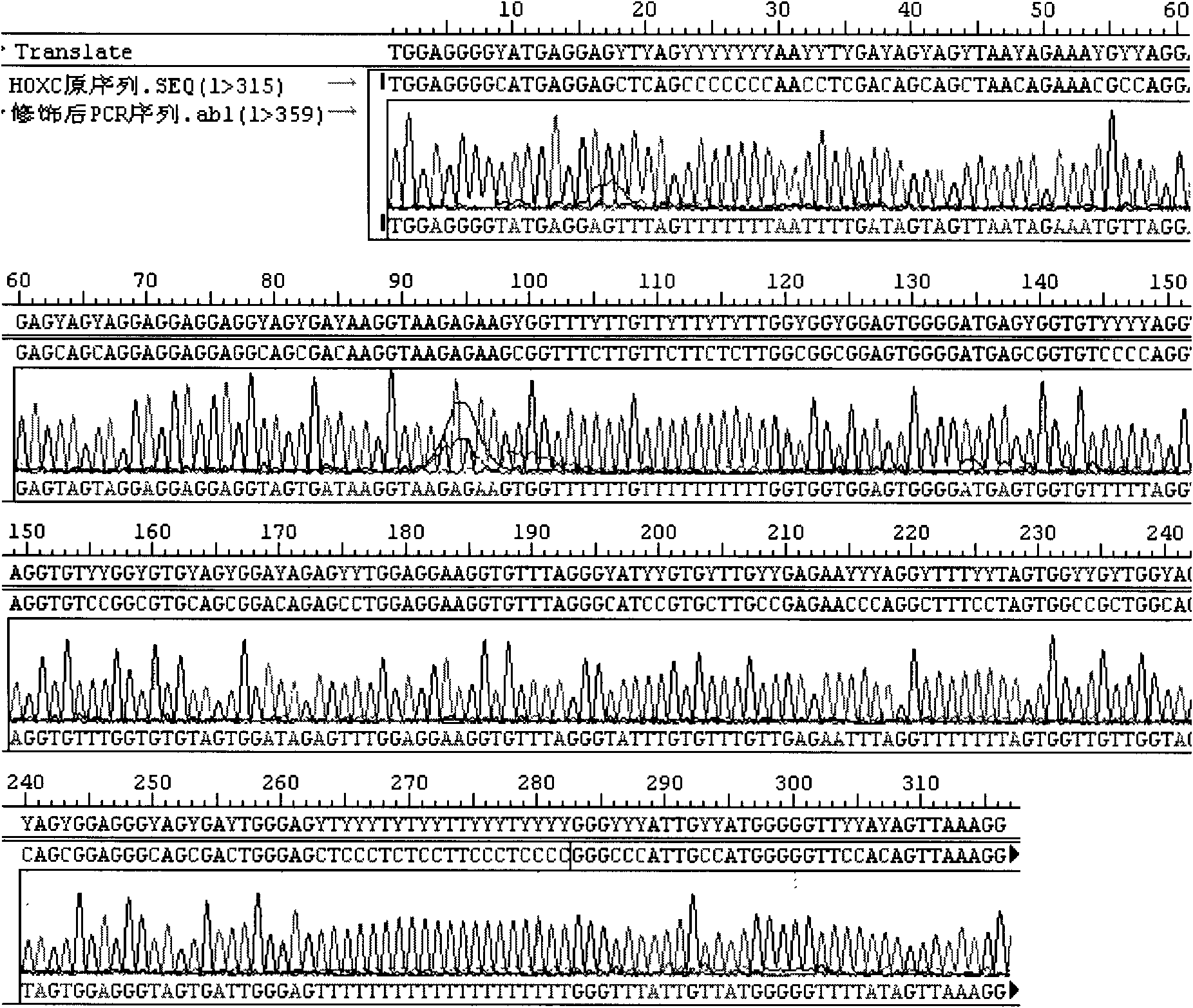
[0086] Example 2: Detection of the HOXC9 gene promoter region in oral epithelial cells
[0087] 1. Sampling: Before sampling, rinse your mouth with clean water, then hold a sterilized medical cotton swab in your hand, insert it into your mouth, wipe it from the buccal mucosa on the inner side of your mouth repeatedly for about 5 minutes, take out the cotton swab, and put the cotton swab into EP containing 1ml TE Wash in the tube, centrifuge at 12,000 rpm for 3 minutes, remove the supernatant, and collect oral epithelial cells.
[0088] 2. Cell lysis: Take 500 ul of the modification solution of formula 1 in Example 1 and put it into a 1.5 ml EP tube, add less than or equal to 20 ul of collected oral epithelial cells into the EP tube filled with the modification solution, mix evenly by inversion, With gentle shaking for 30 seconds, the lysed cells became clear.
[0089] 3. Modification: Divide the above modification liquid into 4 parts and put them into 4 PCR tubes, and perform...
Embodiment 3
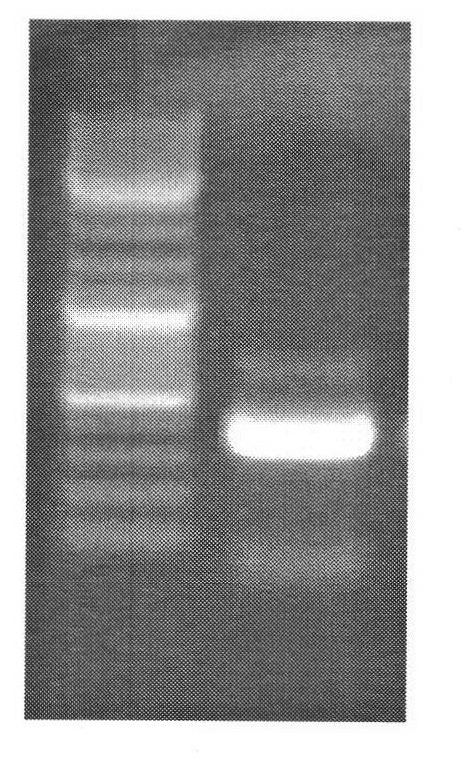
[0107] Example 3: Detection of the CRABP1 gene promoter region associated with tumor metastasis in tumor tissue slices
[0108] Tissue section dewaxing: use sterilized forceps to slice paraffin tissue about 30-100mm 2 Clip it into a 1.5ml EP tube, add 100ul xylene, flick the EP tube, let it stand at room temperature for 3min, then centrifuge at 13000rpm for 5min, and discard the supernatant with a pipette. Add 100ul of absolute ethanol, centrifuge at 12000rpm for 2min, and discard the supernatant with a pipette. Add 100ul of 75% ethanol, centrifuge at 12000rpm for 2min, carefully discard the supernatant, and dry the sample.
[0109] Cell lysis: Take 500 ul of the modified solution of formula 1 in Example 1 and put it into the EP tube containing the sample, mix it upside down, shake slightly for 30 seconds, and the cell lysis becomes clear.
[0110] Follow steps 3-6 in Example 2.
[0111] PCR:
[0112] Upstream primer: 5'TTGAGAAGATGAGGAATGGTGA 3' (SEQ ID NO: 3)
[0113] Do...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com