Method for preparing food grade Saccharomyces cerevisiae recombinant plasmid
A technology of Saccharomyces cerevisiae and recombinant plasmids, which is applied in the field of biological genetic engineering and can solve problems such as hidden dangers of fermentation system pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
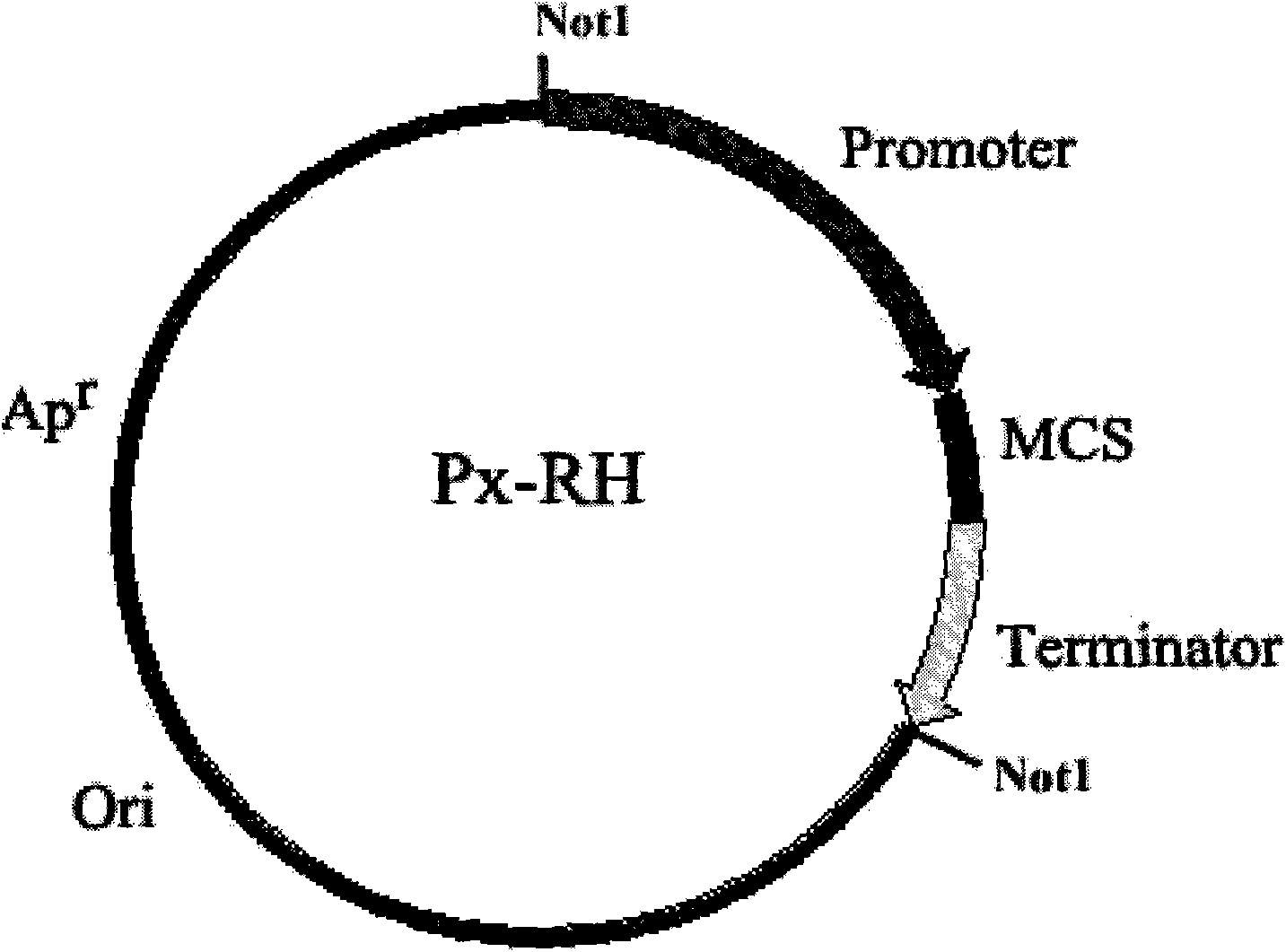
[0054] Example 1: Cloning vector P inu -RH build
[0055] (1) pAG61 plasmid Not1 digestion, electrophoresis recovery Escherichia coli plasmid replicon (Ori)-ampicillin resistance gene (Ap r ) part, alkaline phosphatase treatment to remove phosphorus.
[0056] (2) Using Kluyveromyces cicerisporus CBS4857 genomic DNA as a template, PCR amplification of the inulinase gene with P1 and P2 as primers (GenBank accession number AF178979, sequence such as SEQ.ID.NO 13, Wen T.Q., Cloning and analysis of the inulinase gene from Kluyveromyces cicerisporus CBS4857, World Journal of Microbiology & Biotechnology 19:423-426, 2003), GenBank accession number is AF17897. ) promoter Pinu fragment. Saccharomyces cerevisiae S288c genomic DNA was used as a template, and the terminator Tcyc fragment of CYC1 gene (SGD accession number YJR048W) was amplified by PCR with primers P3 and P4. Carry out corresponding restriction endonuclease digestion treatment according to the enzyme cutting sites desi...
Embodiment 2
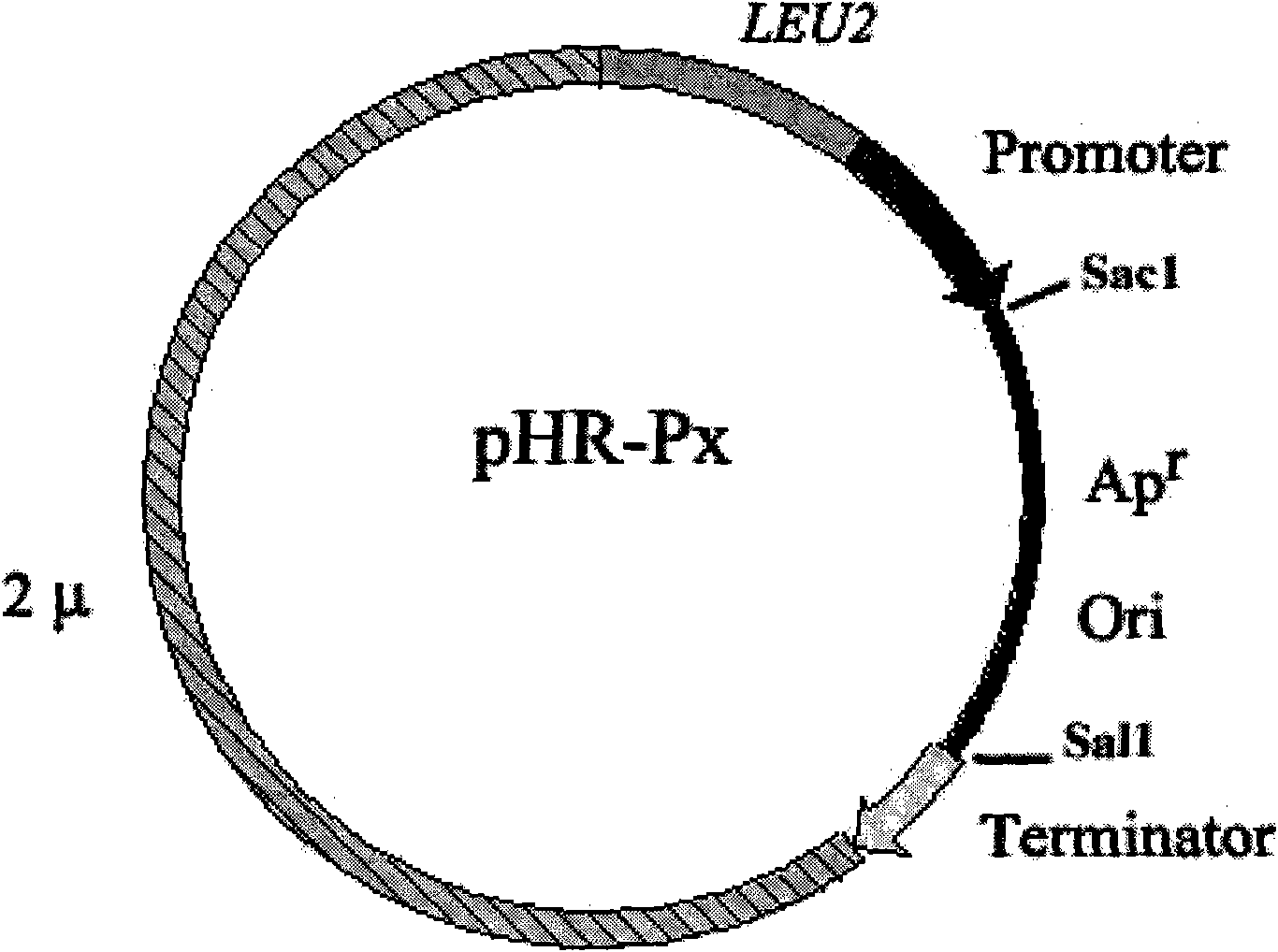
[0070] Example 2: Shuttle carrier pHR-P inu Construct
[0071] (1) The pHC11 vector was digested with Bgl I and Sal double enzymes, and a large fragment of 8.7kb (including the complete 2μ plasmid sequence and Leu2 gene sequence) was recovered, and treated with Klenow enzyme to make up.
[0072] (2) Primers P7 and P8 PCR amplified inulinase gene promoter Pinu, and primers P9 and P10 PCR amplified CYC1 gene terminator Tcyc. The Pinu PCR product was digested with EcoR1 and BamH1, and the Tcyc PCR product was digested with BamH1 and HindIII. The Pinu and Tcyc fragments were ligated with the plasmid pSP72 that had been digested with HindIII and EcoR1, and transformed into Escherichia coli DH5α. And sequence identification, get pSP72-Pinu-Tcyc plasmid.
[0073] (3) The pSP72-Pinu-Tcyc plasmid was linearized with BamH1 single-enzyme digestion, filled in with Klenow enzyme, ligated with the pHC11 enzyme-digested fragment (8.7kb) in (1), transformed into Escherichia coli DH5α, posit...
Embodiment 3
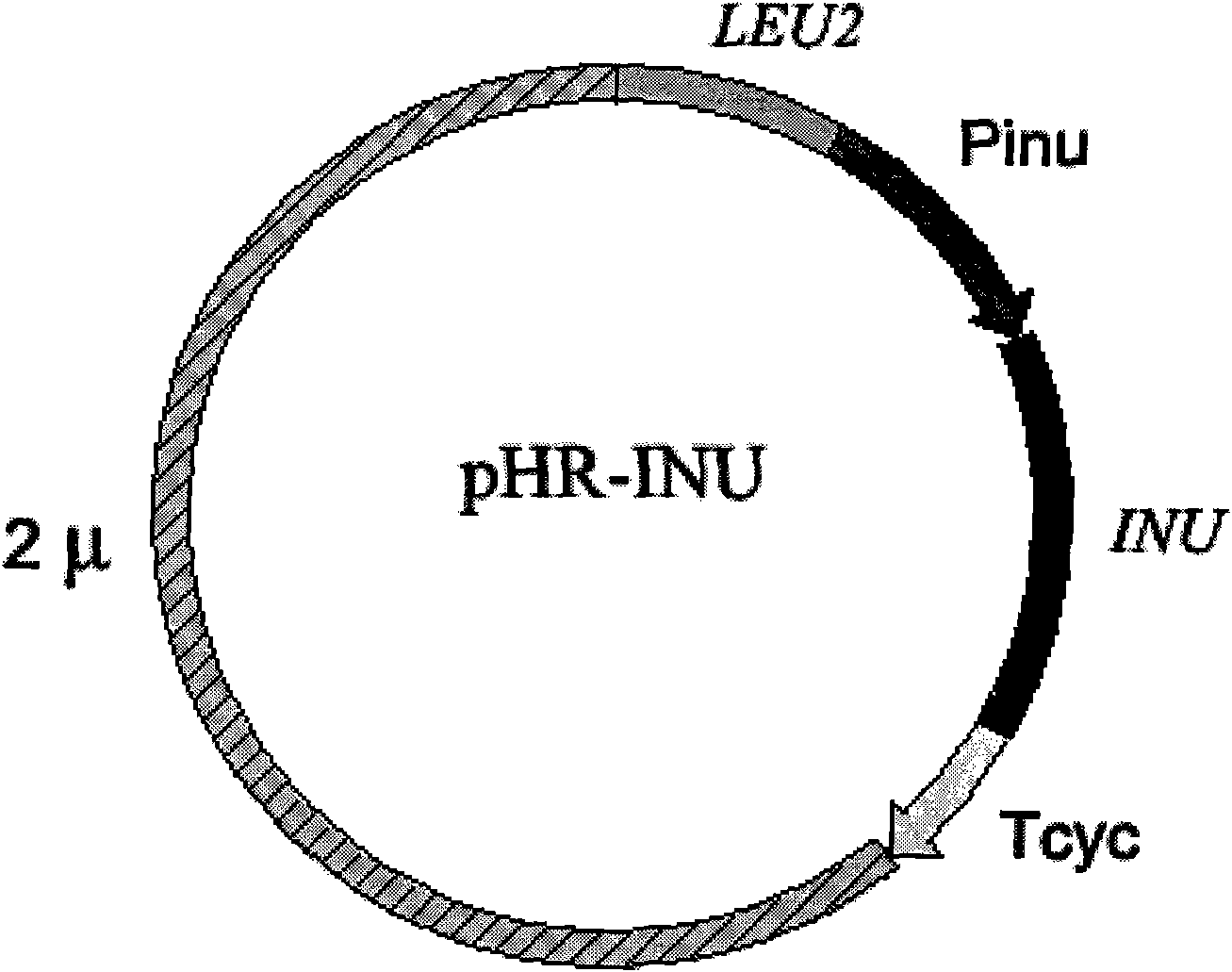
[0085] Embodiment 3: construction of inulinase gene expression cassette
[0086] Kluyveromyces cicerisporus CBS4857 genomic DNA was used as a template, primers P11 and P12 were used to PCR amplify the open reading frame ORF of the inulinase gene, and the PCR product was digested with HindIII and Sal1, and combined with Pinu - RH vector ligated to transform Escherichia coli DH5α, identify the transformant by PCR, extract the recombinant plasmid of the positive transformant, sequence and identify, obtain the recombinant plasmid in which the inulinase gene is cloned in the MCS region of the Pinu-RH vector, and the recombinant plasmid has a Pinu promoter- The inulinase gene expression cassette (Pin-Inu-Tcyc) composed of the inulinase gene-Tcyc terminator, the recombinant plasmid was digested with Not1, separated by electrophoresis, and the inulinase gene expression cassette (Pin-Inu-Tcyc) was recovered.
[0087] Inulinase gene open reading frame ORF cloning primers:
[0088] P11:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com