Method for increasing exogenous gene expression quantity and Chlamydomonas reinhardtii hydrogen production quantity by codon optimization
A technology for optimizing codons and exogenous genes, which can be used in the fields of exogenous gene expression and biological hydrogen production, and can solve problems such as limitation and inactivation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Chlamydomonas reinhardtii culture:
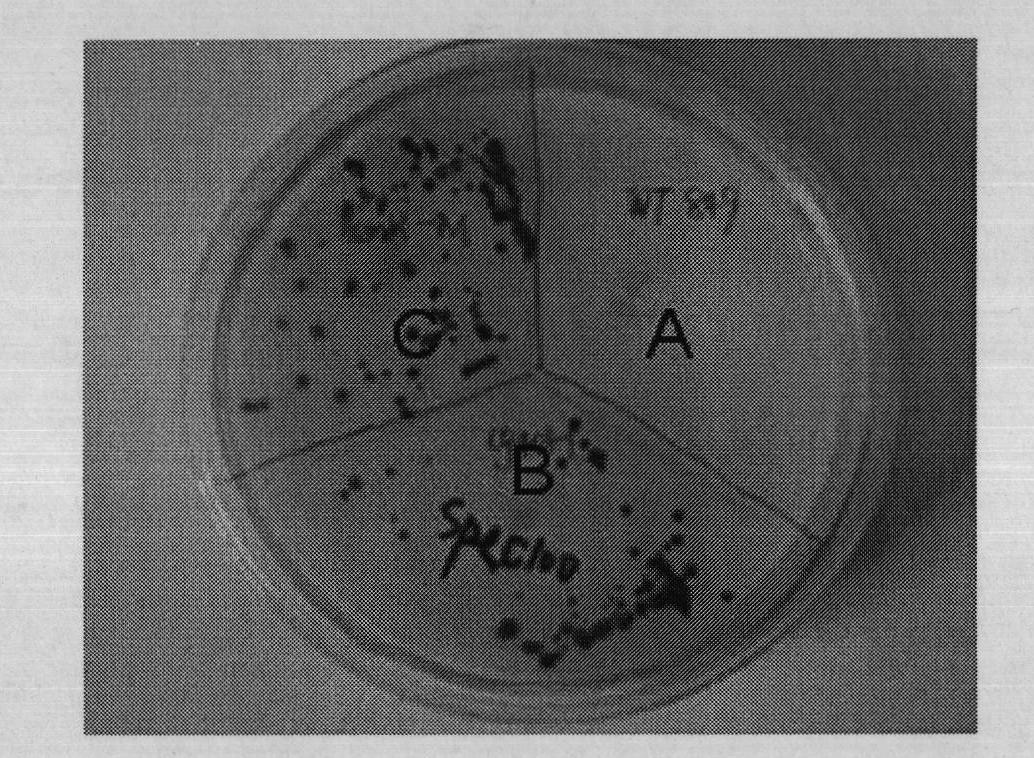
[0091] Chlamydomonas reinhardtii has fast growth rate, low culture cost and high hydrogenase activity, and is a representative species of microalgae for photosynthetic hydrogen production. In order to facilitate transgene manipulation, the present invention prefers Chlamydomonas reinhardtii species cc849 with a cell wall deficiency.
[0092] ①Normal culture conditions of Chlamydomonas reinhardtii: According to "The Chlamydomonas Sourcebook: a comprehensive guide to biology and laboratory use. New York: Academic Press. 1989" edited by Harris, the optimal condition is 25±1°C, and the intensity of sunlight (100-200 μmol photons m -2 the s -1 ); liquid culture is in 50~100ml Tris-Acetate-Phosphate (TAP) medium, initial pH7.2, horizontal shaker speed 100~130rpm, every 5~6 days 1% inoculation subculture; Solid TAP plate culture medium contains 1.5% agar powder, the preservation and purification of the algal species is to pick a single cl...
Embodiment 2
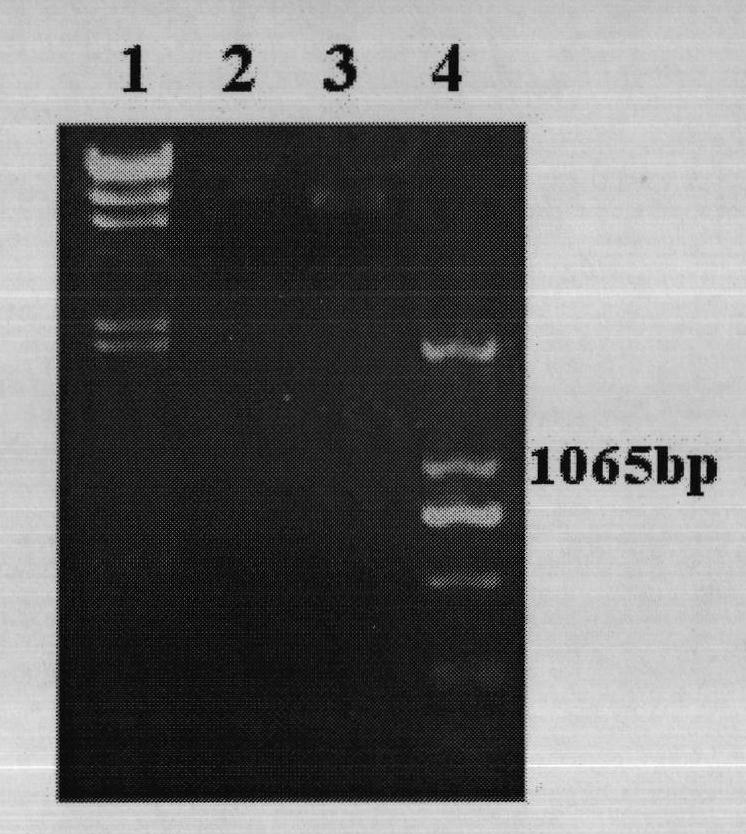
[0096] Codon optimization, synthesis and PCR amplification of leghemoglobin hemH gene and lba gene:
[0097] The Bradyrhizobia hemH gene sequence whose sequence number is M92427 and the soybean lba gene sequence whose sequence number is V00453 are retrieved from GenBank;
[0098] The codons expressed by Chlamydomonas reinhardtii chloroplast genes have obvious adenine (A) and uracil (U) biases, especially the third base of the codons has obvious A and U biases. According to the degeneracy of coding amino acid codons and without changing the original gene hydrogen acid sequence, the codons of leghemoglobin hemH gene and lba gene were respectively modified, and the DNA nucleotide sequence corresponding to the original codons , especially in the DNA nucleotide sequence corresponding to the third base of the codon, the original guanine (G) or cytosine (C) is changed to adenine (A) or thymine (T), as shown in Annex 1 and Shown in Annex 2. The optimized genes were named hemHc and l...
Embodiment 3
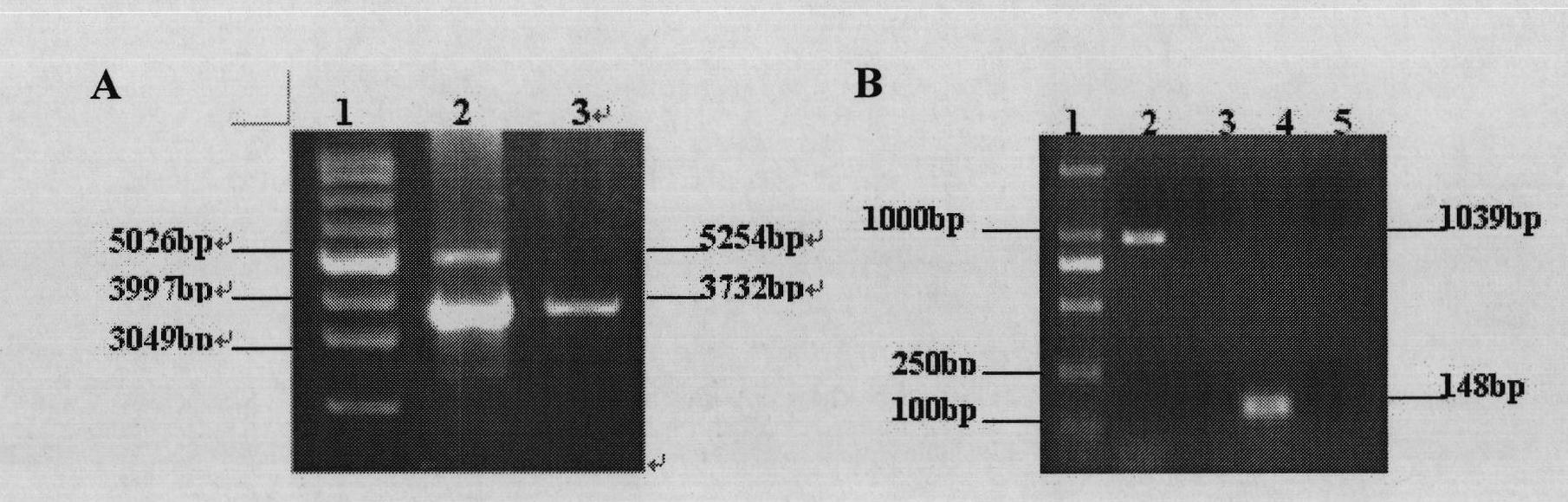
[0110] Construction of Chlamydomonas reinhardtii chloroplast expression vector:
[0111] By standard methods known to those skilled in the art (Sambrook et al., Molecular Cloning. New York: Cold Spring Harbor Laboratory Press. 1998) and the method described by Vaistij et al. for Chlamydomonas reinhardtii chloroplast expression vector cg40 (The Plant Journal, 2000, 21( 5): 469-482) for vector construction. Respectively cut the lbac gene fragment and vector cg401-1 obtained from the above-mentioned clone with restriction endonucleases SmaI and SacI, and connect the purified lbac gene fragment to the plasmid cg401-1 with T4 DNA ligase through the above two enzyme cutting sites After the aadA gene in the vector, the aadA-lbac fusion gene was formed to obtain the vector cg401-1-lbac, and then the hemHc gene was connected to the aadA gene and the lbac gene in the plasmid cg401-1-lbac using the SacII restriction site with T4 DNA ligase Before, the aadA-hemHc-lbac fusion protein was ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com