Method for detecting DNA (deoxyribonucleic acid) content of CHO (cholesterol) cells by probe
A cell and probe technology, applied in the field of quantitative detection of CHO cell DNA content, can solve the problems of high implementation cost and low sensitivity, and achieve the effect of high specificity and good sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Design of primers and preparation of reference materials for the detection of DNA residues in CHO cells by fluorescent quantitative PCR
[0037] 1. Materials and reagents
[0038] DNTPs, Taq DNA polymerase, fluorescent dyes and PCR buffer are commercially available products. The 7500Fast fluorescent quantitative PCR instrument is a product of Applied Biosystems in the United States. The DNA diluent is prepared by our laboratory using analytical reagents. Product of Bio-Tek Corporation.
[0039] 2. Design and synthesis of primers and probes
[0040] Using the Genebank accession sequence (Genebank accession number EF540878.1) as a template, use Primer Primer 5.0 software (Premier Biosoft, USA) to design PCR primers and probes, and select the following combinations from the preliminary experiments.
[0041] The nucleotide sequence of Primer-F is: 5'-ACAGGTTTCTGCTTCTGGCT-3';
[0042] The nucleotide sequence of Primer-R is: 5'-TAGCAGACACTGTTGTAGAG-3';
[0043] Probe: 5'-...
Embodiment 2
[0052] Example 2 Application of Fluorescent Quantitative PCR to Detect CHO Cell DNA Concentration in Analysis of CHO Cell Gene Copy Number
[0053] 1. The purpose is to provide an internal reference for analyzing the gene copy number of the target gene.
[0054] 2. Preparation of CHO cell DNA
[0055] Collect CHO cells of different cell lines and different passage times, use genomic DNA extraction kit to extract genomic DNA, first measure the concentration of extracted genomic DNA by spectrophotometry, and dilute the samples appropriately so that each sample is within the range of the standard curve .
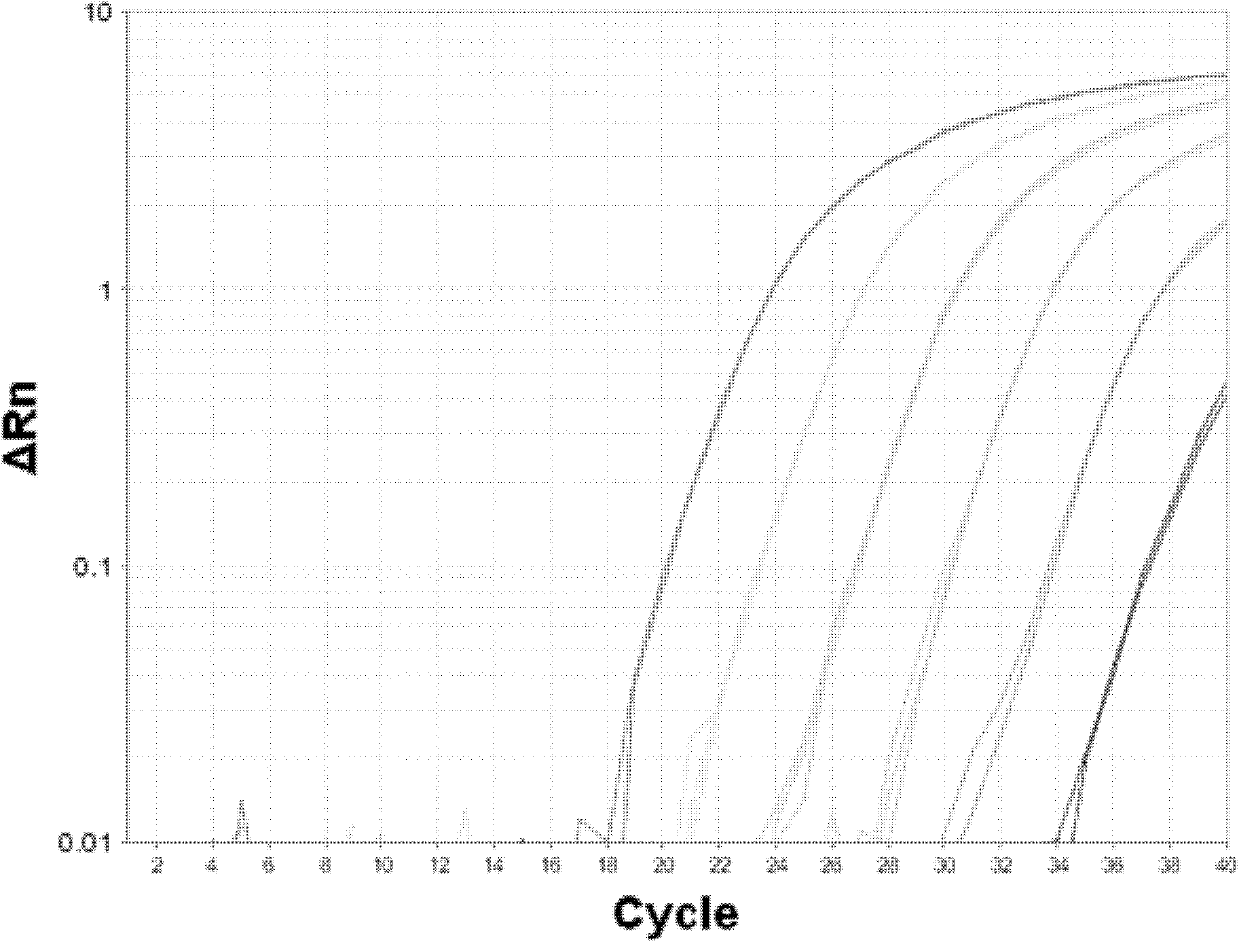
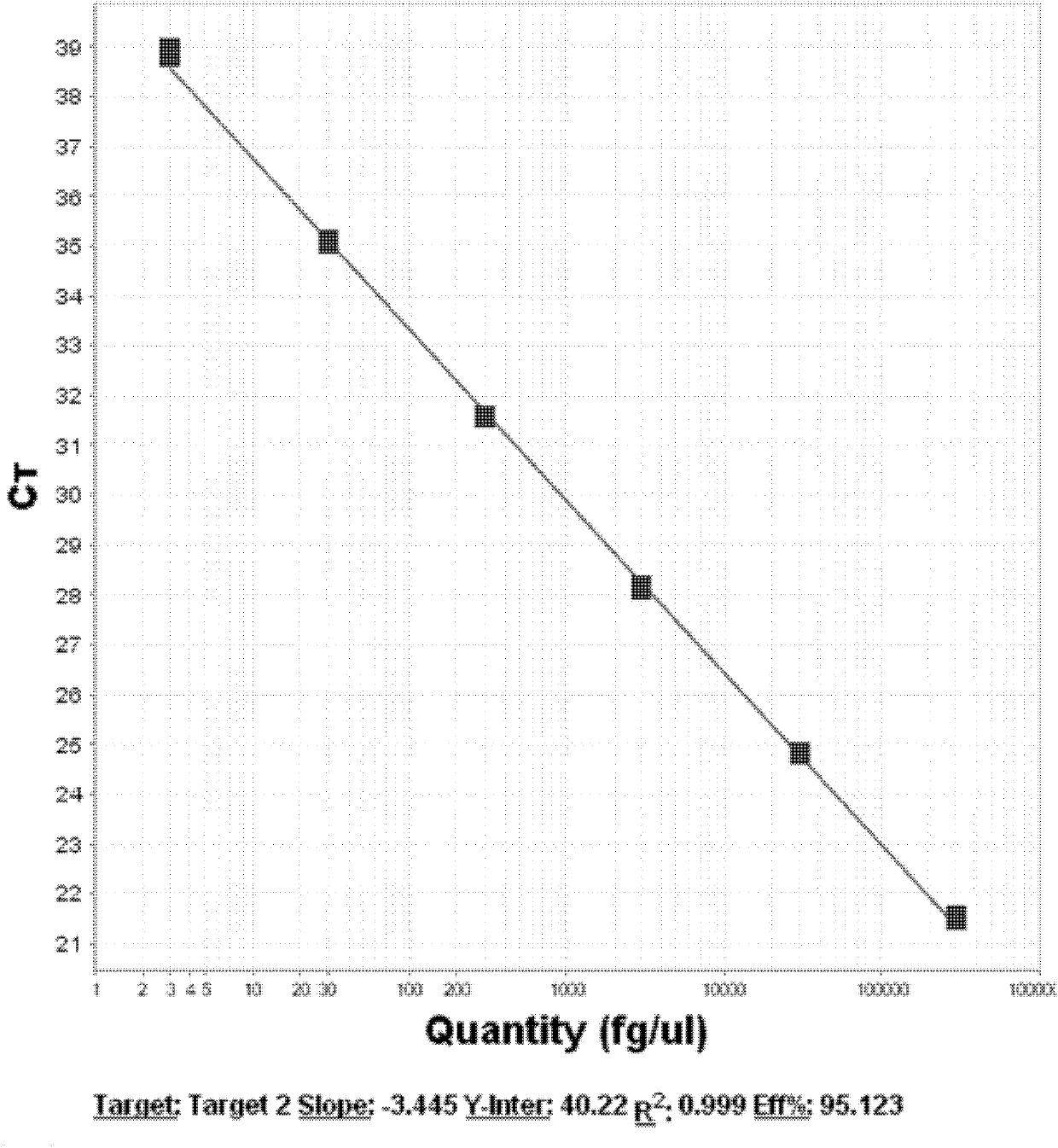
[0056] 3. Select 7 concentrations of reference substances, the concentrations of which are 3.0×106fg / μl, 3.0×105fg / μl, 3.0×104fg / μl, 3.0×103fg / μl, 3.0×102fg / μl, 3.0×101fg / μl , 3.0fg / μl Refer to the method described in Example 1 for quantitative PCR analysis. Amplification results and standard curves such as image 3 , Figure 4 shown.
[0057] 4. The expression of the tar...
Embodiment 3
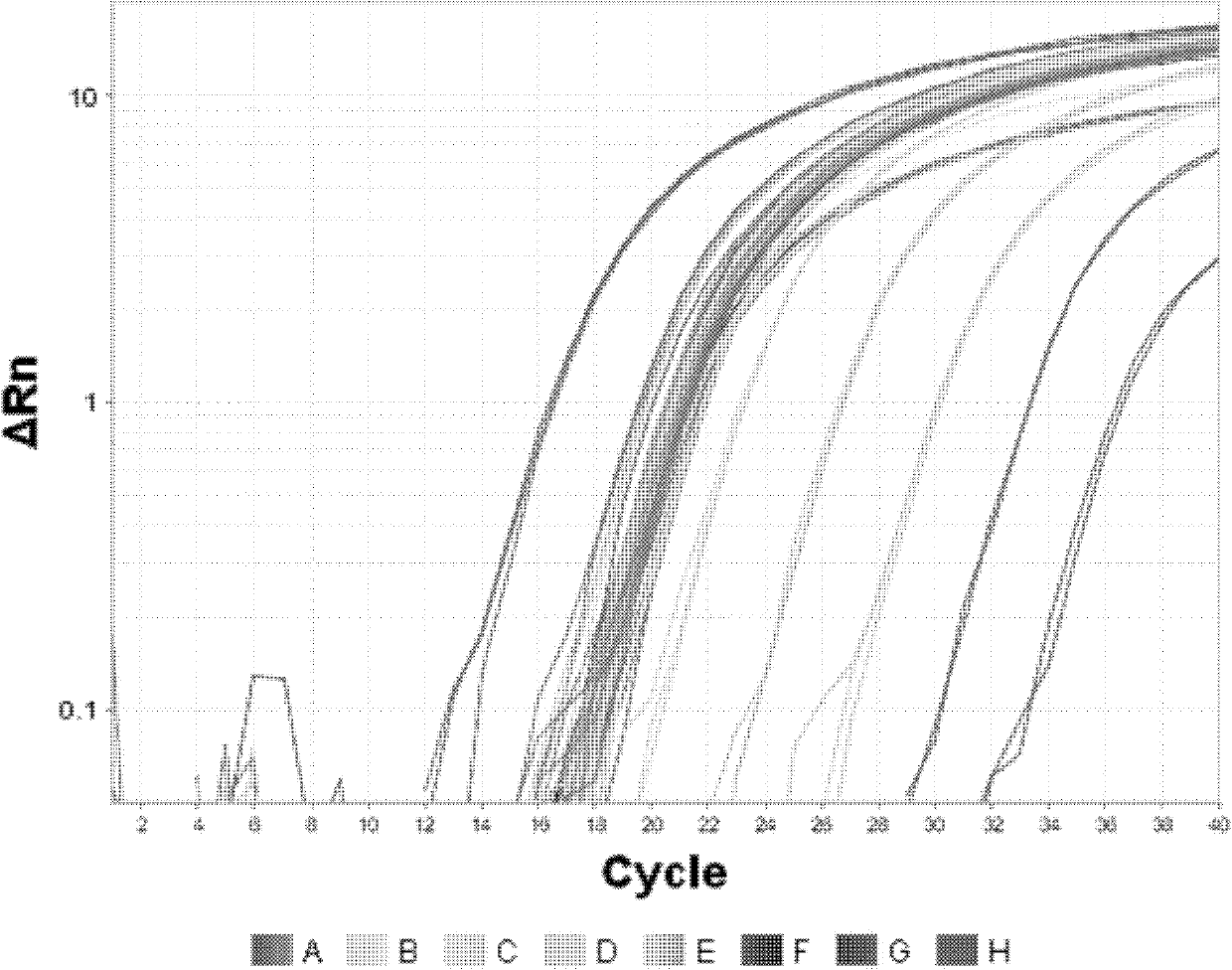
[0058] Example 3 Application and Specificity Analysis of Fluorescent Quantitative PCR Detection of Residual DNA in CHO Cells
[0059] 1. Preparation includes the following PCR amplification reaction solution and dilution solution:
[0060] PCR system: PCR buffer (10X) 3.0μl; dNTP (2mM / L) 2.0μl; MgCl 2 (50mM / L) 2.0μl; Rox dye (50X) 0.6μl; forward primer (10μmol / L) 1.0μl; reverse primer (10μmol / L) 1.0μl; probe (10μmol / L) 1.5μl Taq enzyme ( 50U / μl) 0.1 μl; template 9.5 μl; pure water 9.3 μl.
[0061] DNA diluent: use deionized water as solvent, Tris-HCl concentration is 5.0mM / L, EDTA concentration is 1.0mM / L, adjust pH to 8.0 with NaOH solution.
[0062] 2. Sample collection, transportation and storage
[0063] Sample collection: including semi-finished and finished products of recombinant protein drugs produced by using CHO cells as host cells, which are operated according to the sampling requirements of the National Institute for the Control of Biological Products, sealed an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com