Silkworm fibroin heavy-chain gene mutation sequence and mutation method and application
A heavy chain gene and silk technology, applied in the fields of application, genetic engineering, plant gene improvement, etc., to achieve the effect of increasing expression and purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1: Sequence analysis of silk fibroin heavy chain gene of silkworm
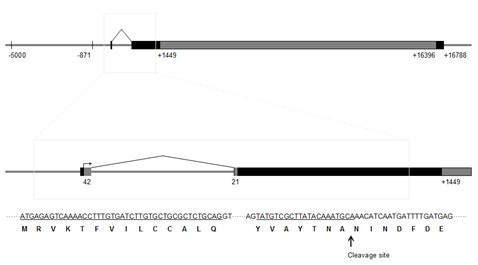
[0055] Download the silk fibroin heavy chain gene sequence (No. AF226688) from the NCBI database, and its sequence structure is as follows: figure 1 shown. Bombyx mori silk fibroin heavy chain gene (+1~+16 788, where +1 indicates the transcription start site) includes two exons with a length of 67bp and 15750bp and an intron with a length of 971bp, the first exon The exon includes a 25bp untranslated region (+1~+25) and a 42bp coding region (+26~+67). region (+16396~+16788) and highly repetitive region (+1450~+16395, figure 1 shown in the gray box). The N-terminal of the amino acid sequence encoded by silkworm silk fibroin heavy chain gene contains a signal peptide with a length of 21 amino acid residues ( figure 1 underlined).
[0056] In order to select zinc finger nuclease target sites that can play a role in various silkworm strains, we performed SNP analysis on the N-terminal part...
Embodiment 2
[0057] Example 2: Design and synthesis of silk fibroin heavy chain gene-specific zinc finger nuclease sequence
[0058] According to the sequence characteristics of silk fibroin heavy chain gene and its SNP distribution in different silkworm strains, combined with the characteristics of zinc finger protein recognition of DNA sequences, we selected CTGTTGCTCAAAGTTATGTTGCTGCTGATGCGGGAGCA as the target site for zinc finger nuclease recognition. +1325-+1362 positions of the sequence shown in SEQ ID NO:1, and design and synthesize zinc finger nuclease accordingly.
[0059] Therefore, positions 1325-1362 of silk fibroin heavy chain gene are the target sites recognized by zinc finger nuclease.
[0060]
Embodiment 3
[0061] Example 3: Preparation of zinc finger nuclease mRNA
[0062] Artificially synthesized or amplified from the existing zinc finger protein library to obtain the nucleic acid sequence of zinc finger nuclease (such as SEQ ID NO: 119 and SEQ ID NO: 120) with restriction endonucleases EcoRI and XhoI (purchased from TAKARA) After double enzyme digestion, carry out a ligation reaction with the prokaryotic expression vector pET28a that has undergone the same enzyme digestion, transform Escherichia coli and screen positive clones to obtain a recombinant vector. The specific reaction system for enzyme digestion is as follows:
[0063]
[0064] The recombinant vector was digested with XhoI and then transcribed in vitro with the MessageMax T7 mRNA in vitro transcription kit (purchased from Epicentre). The reaction system was as follows:
[0065]
[0066] Incubate the above reaction system at 37°C for 30 minutes, then add 1 μl DNase, and continue to incubate for 15 minutes. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com