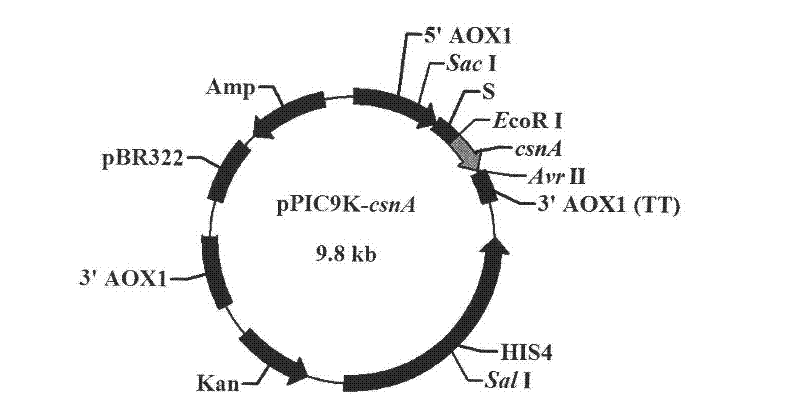
Beta-1, 4-endo-chitosanase (Aus CsnA) gene cloning and preparation of recombinase
A chitosanase and gene technology, applied in the field of bioengineering, can solve problems such as no reports, and achieve the effects of large-scale industrial production, high catalytic activity and thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
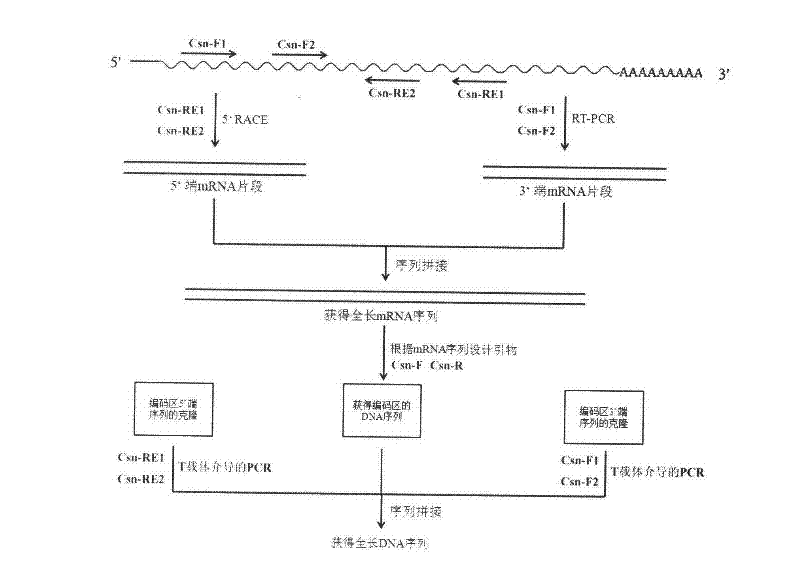
[0035] Cloning of Example 1 Aus csnA 3' end mRNA sequence
[0036] The first strand of cDNA was synthesized by reverse transcription with Oligo dT-Adaptor Primer as primers; the first round of PCR was performed with M13 Primer M4 and Csn-F1 as primers, and the reaction conditions were: 94°C for 2min, 30 cycles (94°C 30s, 52°C for 30s, 72°C for 90s), 72°C for 10min; use M13 Primer M4 and Csn-F2 as primers for the second round of PCR, and the reaction conditions are: 94°C for 2min, 30 cycles (94°C for 30s, 52°C 30s, 72°C 90s), 72°C 10min. The two rounds of PCR products were analyzed by 1% agarose gel electrophoresis, and the target band was recovered and ligated with pUCm-T (pUCm-T-csnA3′), transformed into JM109, and then sent to Shanghai Sangon for sequencing.
Embodiment 2A
[0037] Cloning of embodiment 2 Aus csnA 5' end mRNA sequence
[0038] The first round of PCR was performed using the Outer Primer and Csn-RE1 of the 5′-Full RACE Kit as primers. The reaction conditions were: 94°C for 3min, 30 cycles (94°C for 30s, 55°C for 30s, 72°C for 60s), 72°C 10 min; the second round of PCR was performed using Inner Primer and Csn-RE2 as primers, and the reaction conditions were: 94°C for 3 min, 30 cycles (94°C for 30s, 55°C for 30s, 72°C for 60s), and 72°C for 10 min. The two rounds of PCR products were analyzed by 1% agarose gel electrophoresis, and the target band was recovered and ligated with pUCm-T (pUCm-T-csnA5′), transformed into JM109, and then sent to Shanghai Sangon for sequencing.
Embodiment 3
[0039] Example 3 Cloning of Aus csnA 5' end promoter sequence
[0040] Using the processed A.usamii E001 genomic DNA as a template, the first round of PCR used T-PrimerF and Csn-RE1 as primers, and the reaction conditions were: 94°C for 4min, 30 cycles (94°C for 30s, 55°C for 30s, 72°C 60s), 72°C for 10min; the second round of PCR uses T-PrimerF and Csn-RE2 as primers, and the reaction conditions are: 94°C, 4min, 30 cycles (94°C for 30s, 55°C for 30s, 72°C for 60s), 72°C 10min. The two rounds of PCR products were analyzed by 1% agarose gel electrophoresis, and the target band was recovered by tapping the gel and ligated with pUCm-T (pUCm-T-csnAP), transformed into JM109, and then sent to Shanghai Sangon for sequencing.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com