Method for extracting total DNA (deoxyribonucleic acid) of mulberry rhizosphere soil microorganisms by adopting plurality of measures
A technology of soil microbes, mulberries, applied in the field of molecular ecology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
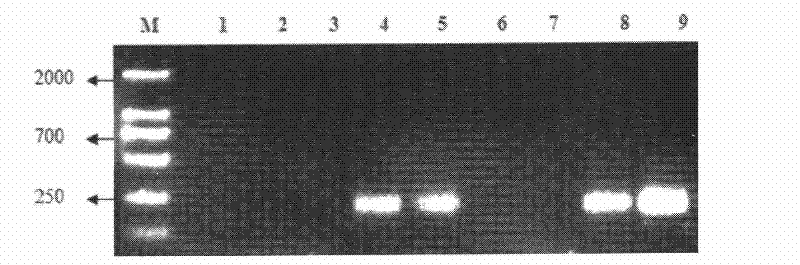
[0052] PVP pretreatment-CTAB-lysozyme-protease-SDS-repeated freeze-thaw method. Take 5g of soil sample and add 30mL of 20g / L sodium metaphosphate buffer solution (containing 10g / L PVP-K30; pH 8.5), oscillate on a shaking table for 15min, centrifuge at 8500r / min for 5min, take the precipitate, and repeat washing 3 times; take the precipitate, add 13.5mL DNA extraction solution I and a little quartz sand, vortex shaker for 3min; add 100mg / mL lysozyme 150μL, mix well, incubate at 37℃, 230r / min shaker for 1h; then add 20mg / mL proteinase K15μL, mix well , in a water bath at 37°C for 1h; add 1.5mL 10% SDS, and bathe in a water bath at 65°C for 1h; :24:1) extraction, 8500r / min centrifugation for 10min; take the supernatant, add an equal volume of chloroform / isoamyl alcohol (volume ratio 24:1) to extract again, 8500r / min centrifugation for 10min, take the supernatant, Add an equal volume of chloroform / isoamyl alcohol (volume ratio 24:1) to extract again, centrifuge at 8500r / min for 1...
Embodiment 2
[0053] PVP pretreatment-CTAB, CaCl2, BSA-lysozyme-protease-SDS-repeated freeze-thaw method Take 5g soil sample and add 20g / L sodium metaphosphate buffer (containing 10g / L PVP-K30; pH 8.5) 30mL, shaker Shake for 15 minutes, centrifuge at 8500r / min for 5 minutes, take the precipitate, and repeat washing 3 times; take the precipitate, add 13.5mL DNA extraction solution II and a little quartz sand, vortex shaker for 3min; add 100mg / mL lysozyme 150μL, mix well, 37 ℃, 230r / min shaker incubate for 1h; then add 20mg / mL proteinase K15μL, mix well, 37℃ water bath for 1h; add 1.5mL 10% SDS, 65℃ water bath for 1h; freeze-thaw 3 times, 8500r / min centrifuge 10min Get the supernatant, add an equal volume of phenol / chloroform / isoamyl alcohol (volume ratio 25:24:1) to extract, and centrifuge at 8500r / min for 10min; get the supernatant, add an equal volume of chloroform / isoamyl alcohol ( The volume ratio is 24:1) and extract again, centrifuge at 8500r / min for 10min, take the supernatant, add 0....
Embodiment 3
[0055] CTAB, PVP, CaCl2, BSA-lysozyme, protease-SDS-repeated freeze-thaw method. Take 5g soil sample, add 13.5mL DNA extraction solution III and a little quartz sand, vortex shaker for 3min; add 150μL 100mg / mL lysozyme, 15μL 20mg / mL proteinase K, mix well, incubate for 1h at 37℃, 230r / min shaker Add 15 μL of 20 mg / mL proteinase K, mix well, and bathe in water at 37°C for 1 hour; add 1.5 mL of 10% SDS, bathe in water at 65°C for 1 hour; Extract with phenol / chloroform / isoamyl alcohol (volume ratio 25:24:1), centrifuge at 8500r / min for 10 min; take the supernatant, add an equal volume of chloroform / isoamyl alcohol (volume ratio 24:1) to extract again, Centrifuge at 8500r / min for 10min, take the supernatant, add 0.7 times the volume of isopropanol and 0.1 times the volume of NaAc to the supernatant, place it overnight at -20°C, centrifuge at 8500r / min, 4°C for 20min, wash the precipitate with 70% ethanol, Centrifuge at 12500r / min for 10min, dry the precipitate at room temperature...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com