Reverse dot-blot hybridization gene chip and production method thereof
A gene chip and bacteria technology, applied in the field of reverse dot hybridization gene chip and its manufacturing, can solve problems such as limiting clinical application value, and achieve the effects of shortening hybridization time, accurate diagnosis and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
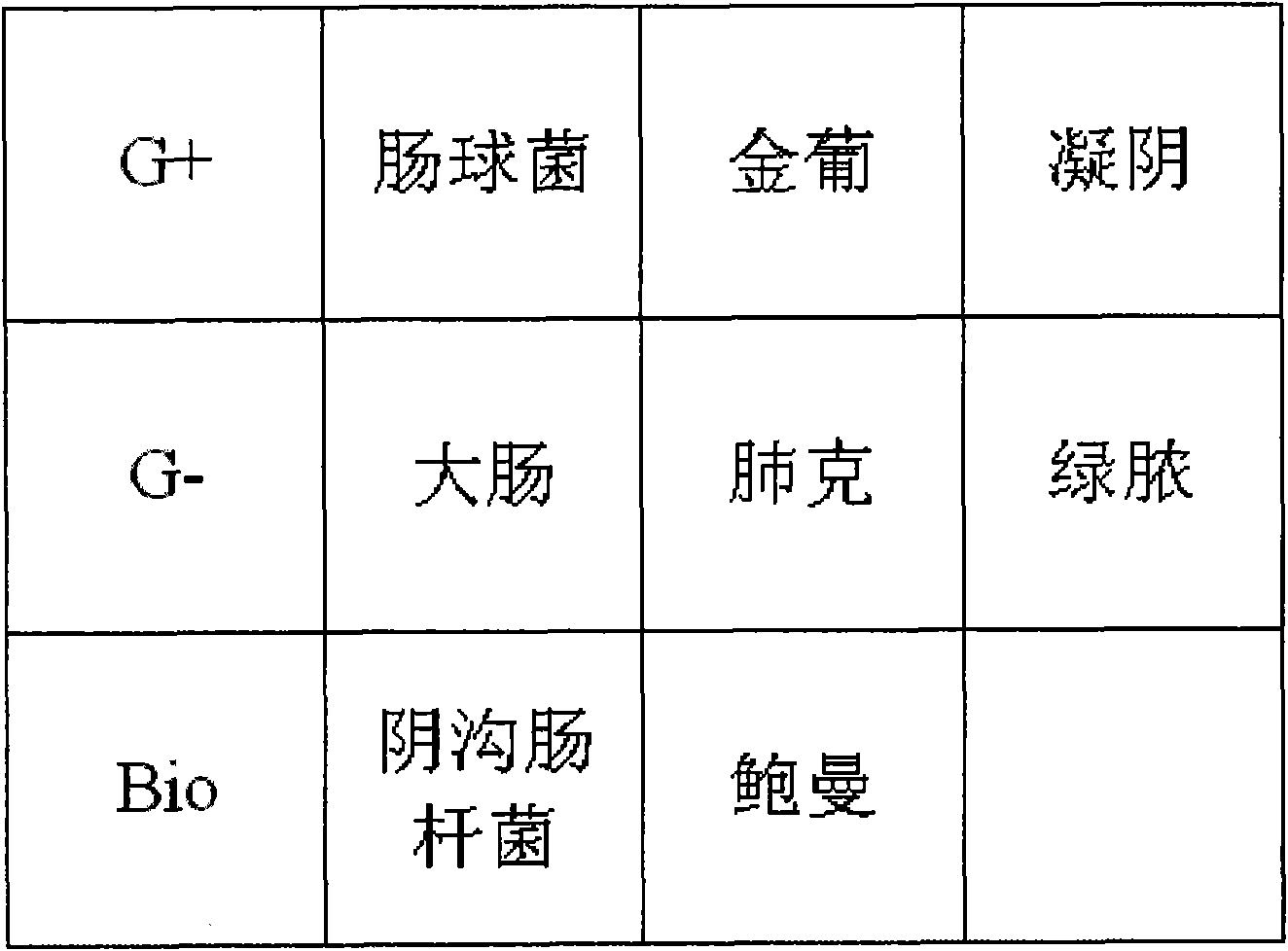
[0049] see figure 1 , a gene chip for rapid screening of pathogenic bacteria in the cerebrospinal fluid of patients after craniotomy, including the general probe for gram-positive bacteria (G+) and the general probe for gram-negative bacteria (G-) set on the matrix , and identify bacterial species, genus-specific probes.
[0050] According to the isolation rate of positive bacteria in cerebrospinal fluid culture and the severity of clinical infection, the types of bacteria identified were selected. The 16s and 23s ribosomal RNA genes of bacteria were selected as target sequences, and species, genus-specific and general sequences were selected from them to design probes for the detection of pathogenic bacteria. The probes of SEQ ID No: 1-10, the nucleotide sequence of each probe is as follows:
[0051] Enterococcus
5'CAAACGAACTTGGAGATAG 3'
SEQ ID No.1
Staphylococcus aureus
5′ CAATCGAACCTGGAGATA 3′
SEQ ID No.2
coagulase-negative staphyloc...
Embodiment 2
[0059] A preparation method of the aforementioned gene chip, according to the isolation rate of clinically positive bacteria in cerebrospinal fluid culture and the severity of clinical infection, the type of bacteria selected for identification. For each bacterial species, design and synthesize specific gene probes with amine groups at 5', and prepare Bio for detection of pathogenic bacteria. This preparation method mainly comprises the steps:
[0060] Preparation of DNA probes: Synthesize the 5' end aminated DNA fragment complementary to the DNA of the strain to be tested, select a specific sequence from the bacterial 16S or 23S ribosomal RNA gene region, and then according to the characteristics of base complementarity, Design and synthesize DNA fragments with amination at the 5' end;
[0061] Immobilization of probes: fix the above probes on the activated nylon membrane, first activate the nylon membrane, then point the probes on the membrane, and form a covalent bond betw...
Embodiment 3
[0075] Preparation of positive control samples
[0076] 1. Use TIANGEN-Bacterial Genomic DNA Extraction Kit to extract positive control bacterial genomic DNA
[0077] a) Take 5ml of Staphylococcus aureus (ATCC25923) bacterium solution prepared with normal saline, the concentration is 103-104;
[0078] b) The bacterial solution was centrifuged at 12000 r / min for 10 minutes, and the supernatant was discarded.
[0079] c) Add 200 μl of 20 mg / ml lysozyme, mix well, and bathe in water at 37° C. for 30 minutes.
[0080] d) Add 20 μl proteinase K and mix well.
[0081] e) Add 220 μl solution B and shake for 15 seconds.
[0082] f) 70°C metal bath for 10 minutes.
[0083] g) Add 220 μl of absolute ethanol and shake for 15 seconds.
[0084] h) Transfer the obtained solution to a clean gel column, centrifuge at 12000 r / min for 30 seconds, and discard the waste liquid.
[0085] i) Add 500 μl of solution D, centrifuge at 12000 r / min for 30 seconds, and discard the waste liquid.
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com