Fusion protein and fusion protein expression vector thereof
A fusion protein and expression vector technology, applied in the field of fusion proteins, can solve the problems of difficulty in obtaining soluble active protein, difficulty in preparation, and high cost, and achieve the effects of facilitating high-density fermentation and culture, low nutritional requirements, and high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1 : Synthesis of genes and primers
[0100] 1. Synthesis of 1exCAR and IgG1 Fc gene
[0101] According to the human-derived CAR extracellular region gene coding sequence (exCAR) (AccessionNumber: NM_001338) and IgG1 Fc segment gene coding sequence (Accession Number: AF237583) in GenBank, exCAR and IgG1 Fc segment were separated according to the partial tropism of Pichia pastoris genetic code The gene sequence was codon-optimized, and Shanghai Sangon synthesized exCAR and IgG1 Fc genes respectively and constructed the engineering bacteria PUC-exCAR / DH 5α , PUC-Fc / DH 5α , the nucleic acid sequence after codon optimization is as follows:
[0102] exCAR (651bp): SEQ ID NO: 1
[0103] ttgtccatcactactccagaagatgattgagaaggctaagggtgagactgcctacttgccatgtaagttcactttgtctccagaagaccaaggtccattggacatcga
[0104] gtggttgatttccccagctgacaatcagaaggttgatcaagtcattattttgtactctggtgacaagatttacgacgactactacccagacttgaagggtagaggttcacttc
[0105] acctccaatgacttgaagtctggtgatgcttctatcaat...
Embodiment 2
[0148] Example 2 : Recombinant engineered bacteria pPIC3.5K-exCAR / DH 5α construction and identification of
[0149] 2.1 exCAR gene EcoR I / SnaB I digestion
[0150] The PUC-exCAR / DH constructed in Example 1.2 5α Bacteria were inoculated in LB (AMP r ) medium, cultivate overnight at 37°C and 180 rpm, extract the PUC-exCAR plasmid with a plasmid extraction kit, and perform step-by-step enzyme digestion with EcoR I and SnaB I.
[0151] ①EcoR I digestion reaction system is as follows:
[0152]
[0153]
[0154] Place at 37°C, and after 3 hours of reaction, directly add SnaB I and corresponding buffer to the system for the second step of digestion.
[0155] ②SnaB I digestion reaction system is as follows:
[0156]
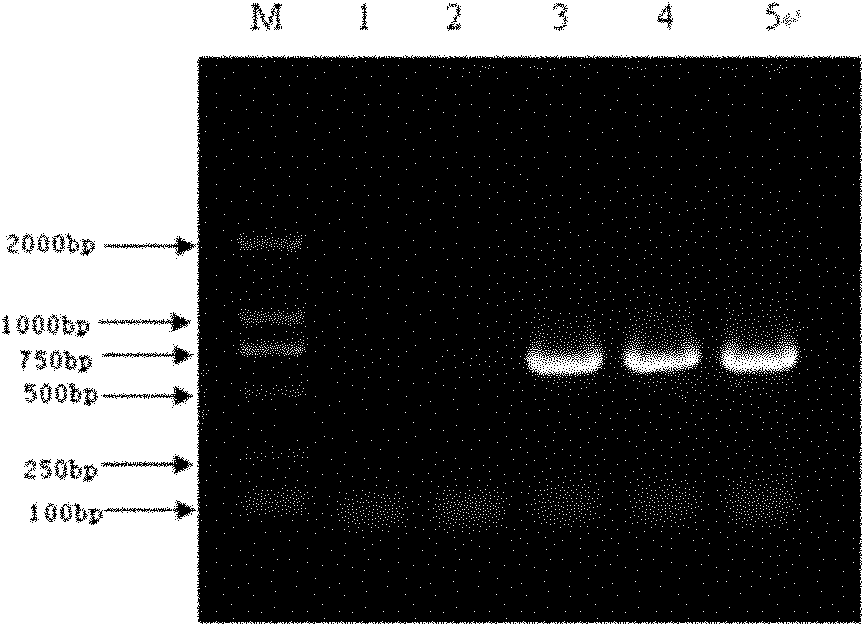
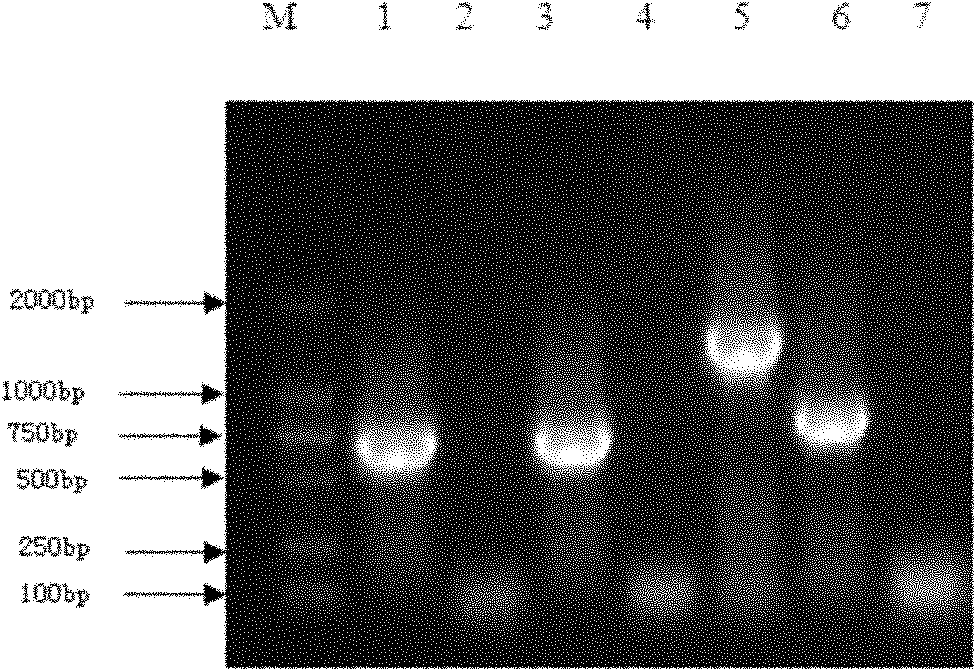
[0157] Set at 37°C and react for 3 hours. The digested product was electrophoresed in 1% agarose gel to recover the exCAR digested gene fragment.
[0158] 2.2 pPIC3.5K plasmid EcoR I / SnaB I digestion
[0159] The plasmid extraction kit extracts the pPIC...
Embodiment 3
[0171] Example 3 : Recombinant engineering bacteria pPIC3.5K-exCAR:Fc / DH 5α build
[0172] 3.1 pPIC3.5K-exCAR plasmid EcoR / Not I double digestion
[0173] The recombinant engineered bacteria plasmid pPIC3.5K-exCAR identified as positive by PCR in Example 2.6 was extracted using a plasmid extraction kit, and subjected to EcoR I / Not I double digestion.
[0174] The enzyme digestion system is as follows:
[0175] Place at 37°C and react for 3 hours. The digested product was electrophoresed in 1% agarose gel, and the digested plasmid pPIC3.5K-exCAR was recovered.
[0176] 3.2 Fc gene Not I / EcoR I double digestion
[0177] The PUC-Fc / DH constructed in Example 1.2 5α Bacteria were inoculated in LB (AMP r ) medium, cultivate overnight at 37°C and 180rpm, extract the PUC-Fc plasmid with a plasmid extraction kit, and perform Not I / EcoR I double enzyme digestion. The reaction system is as follows:
[0178]
[0179] Place at 37°C and react for 3 hours. The digested product w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com