Reconstructed nucleic acid construction and method for quickly recycling amino acid composition by using reconstructed nucleic acid construction
A technology for constructs and amino acids, which is applied in the field of recombinant nucleic acid constructs and the rapid recovery of amino acid compositions using the recombinant nucleic acid constructs, can solve the problems that cells are not easy to amplify and culture, leak, and limit applicability, etc. Achieve the effect of improving purification efficiency, reducing investment and simplifying processing procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
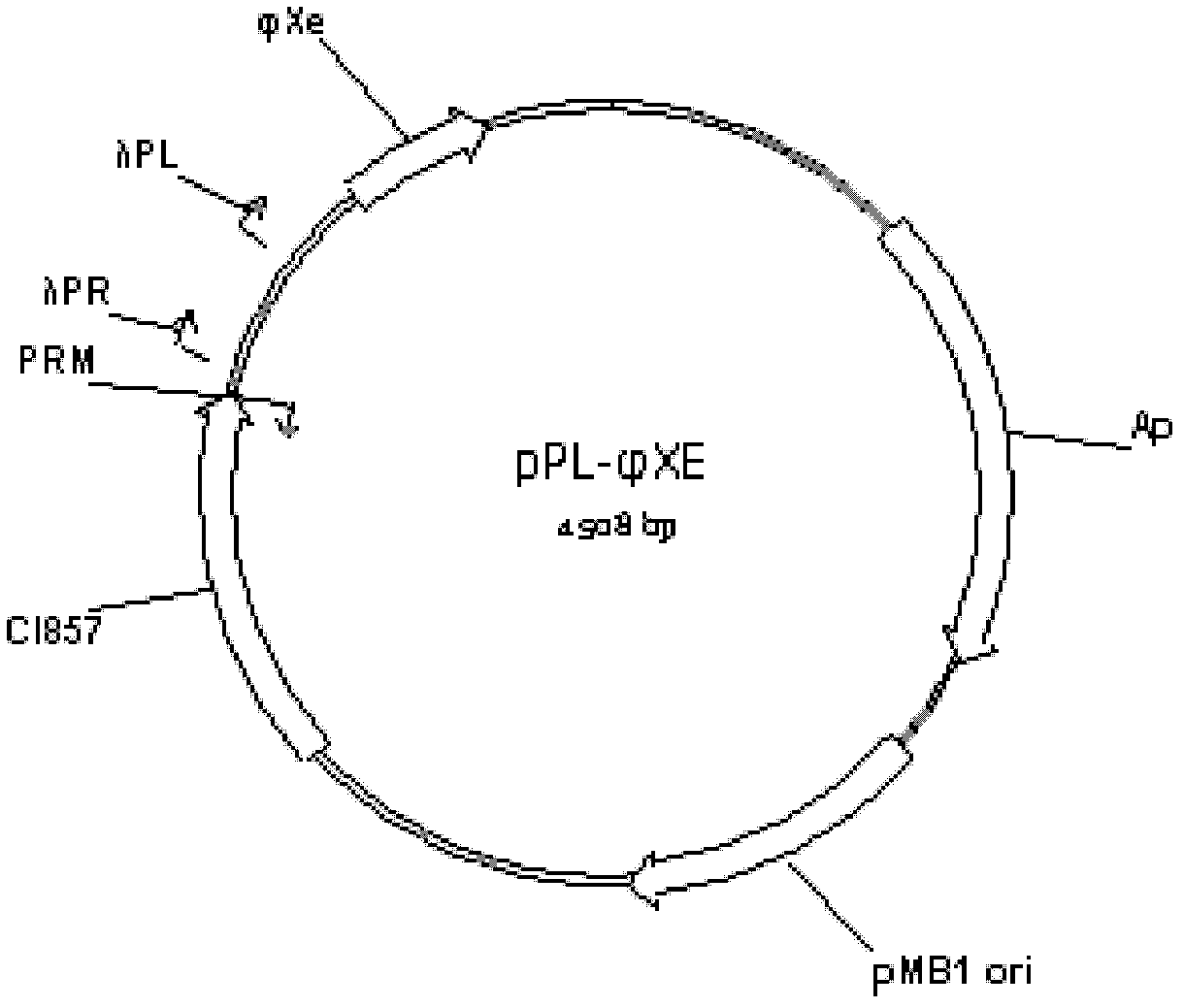
[0032] Example 1: Construction of temperature-induced recombinant nucleic acid construct pPL-Φxe
[0033] temperature-inducible recombinant nucleic acid construct Contains λP R P L The bacteriophage φX174 lytic gene E (φXe) driven by the promoter (promoter) expression, multiple cloning sites, receptor P RM Promoter-regulated heat-sensitive repressor protein (heat-sensitive repressor protein) CI857 gene, pMB1 plastid replication origin from Escherichia coli, and ampicillin-resistant gene. The construction process is as follows. Firstly, the φXe gene primer is synthesized according to the nucleotide sequence of φXe (Refseq: NC 001422) in the genome database of the National Center for Biotechnology Information (NCBI) in the United States:
[0034] Forward introduction:
[0035] (5'TTGCG CATATG GTACGCTGGACTTGTG3')
[0036] Reverse Primer:
[0037] (5'TTTT GAATTC AGACATTTTATTCACTCCTTCCGCACG3')
[0038] The forward primer was designed to contain the cleavage site of rest...
Embodiment 2
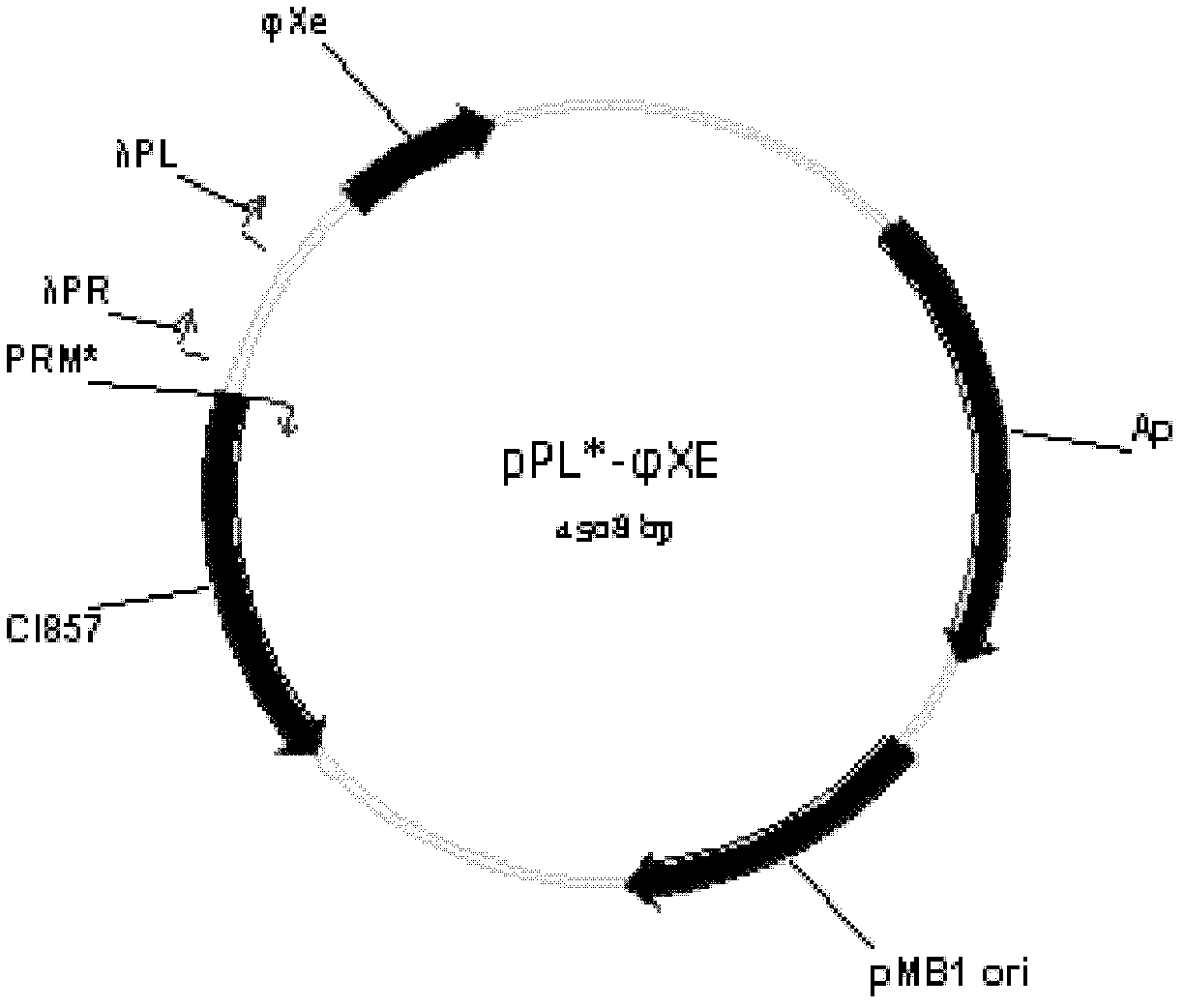
[0039] Example 2: Construction of a temperature-induced recombinant nucleic acid construct
[0040] temperature-inducible recombinant nucleic acid construct Contains λP R P L Promoter-driven expression of φXe, diverse clonal positions, regulated by mutant P RM Promoter (P RM *) Regulated thermosensitive inhibitory protein CI857 gene, pMB1 plastid replication region from E. coli, ampicillin resistance gene. According to published literature (Jechlinger W. et al., 2005, J.Biotechnol.116: 11-20), P RM * Promoter strength is higher than that of P RM Promoter enhancement, thus increasing the expression of the heat-sensitive inhibitory protein CI857 gene, to effectively repress λP R P L Expression of the promoter in the uninduced state. The construction process is as follows. Firstly, point mutation primers were designed according to the pPL452 sequence (GenBank: AB248920.1) of the Genome Database of the National Center for Biotechnology Information. The primer sequence...
Embodiment 3
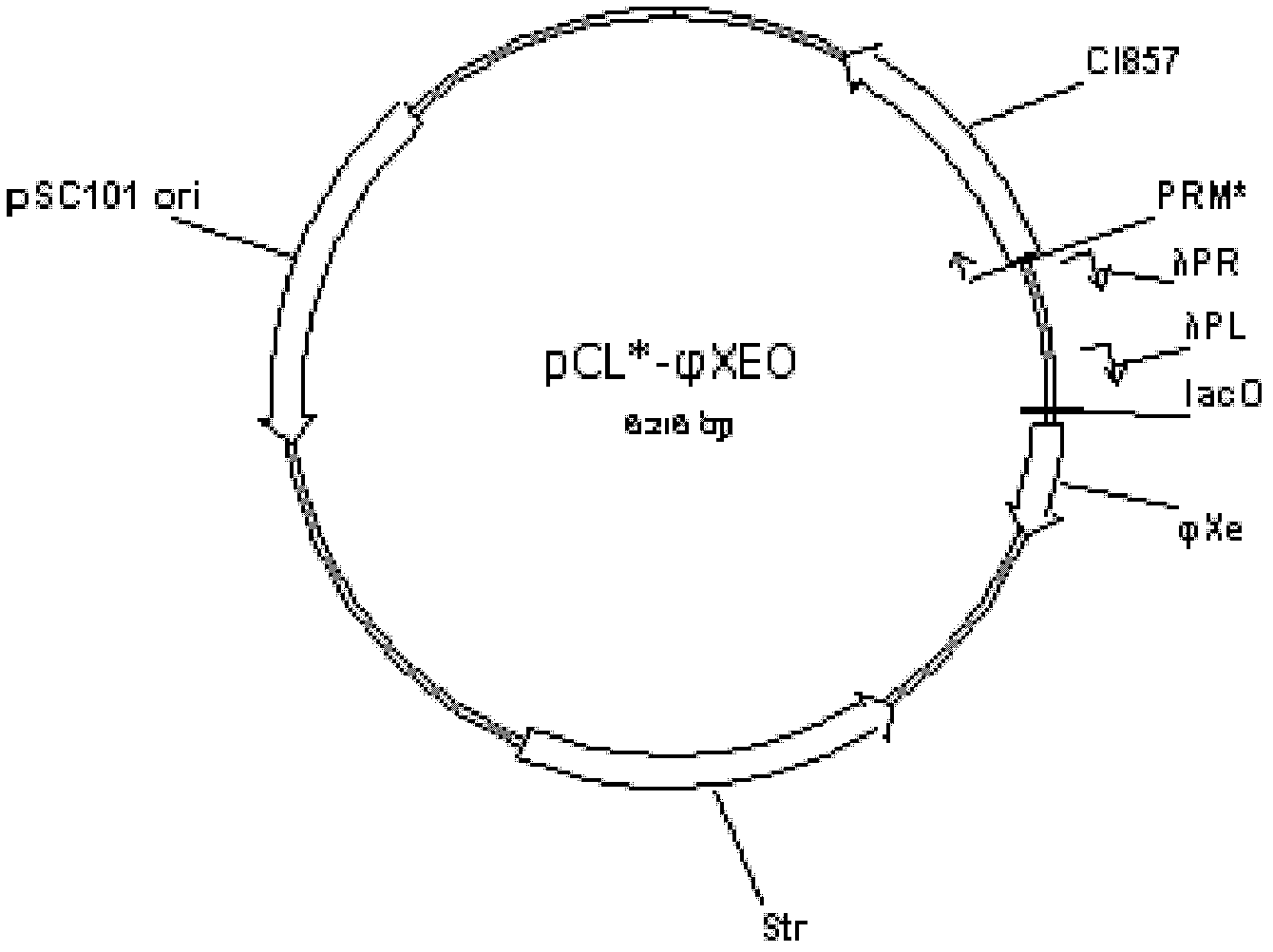
[0046] Example 3: Construction of a temperature-induced recombinant nucleic acid construct
[0047] temperature-inducible recombinant nucleic acid construct Contains λP R P L Promoter-driven expression of φXe and phage (phage) T4 lysin gene (Gpe), regulated by P RM * Regulated thermosensitive inhibitory protein CI857 gene, a lacO operator site, pSC101 plastid replication region, streptomycin resistance gene. The construction process is as follows. First, the primer sequence is designed according to the position of the lacO control subunit in the genome database of the National Center for Biotechnology Information:
[0048] Forward introduction:
[0049] (5' GGATAACAATT GATCCTAAGGAGGTTGATCC3')
[0050] Reverse Primer:
[0051] (5' GCTCACAATT CCAATGCTTCGTTTCGTATCACACACC3')
[0052] The above primer pair contains a lacO control sub-position (as indicated by the underline), and the plastid constructed in Example 2 As a DNA template, use PCR amplification reaction, us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com