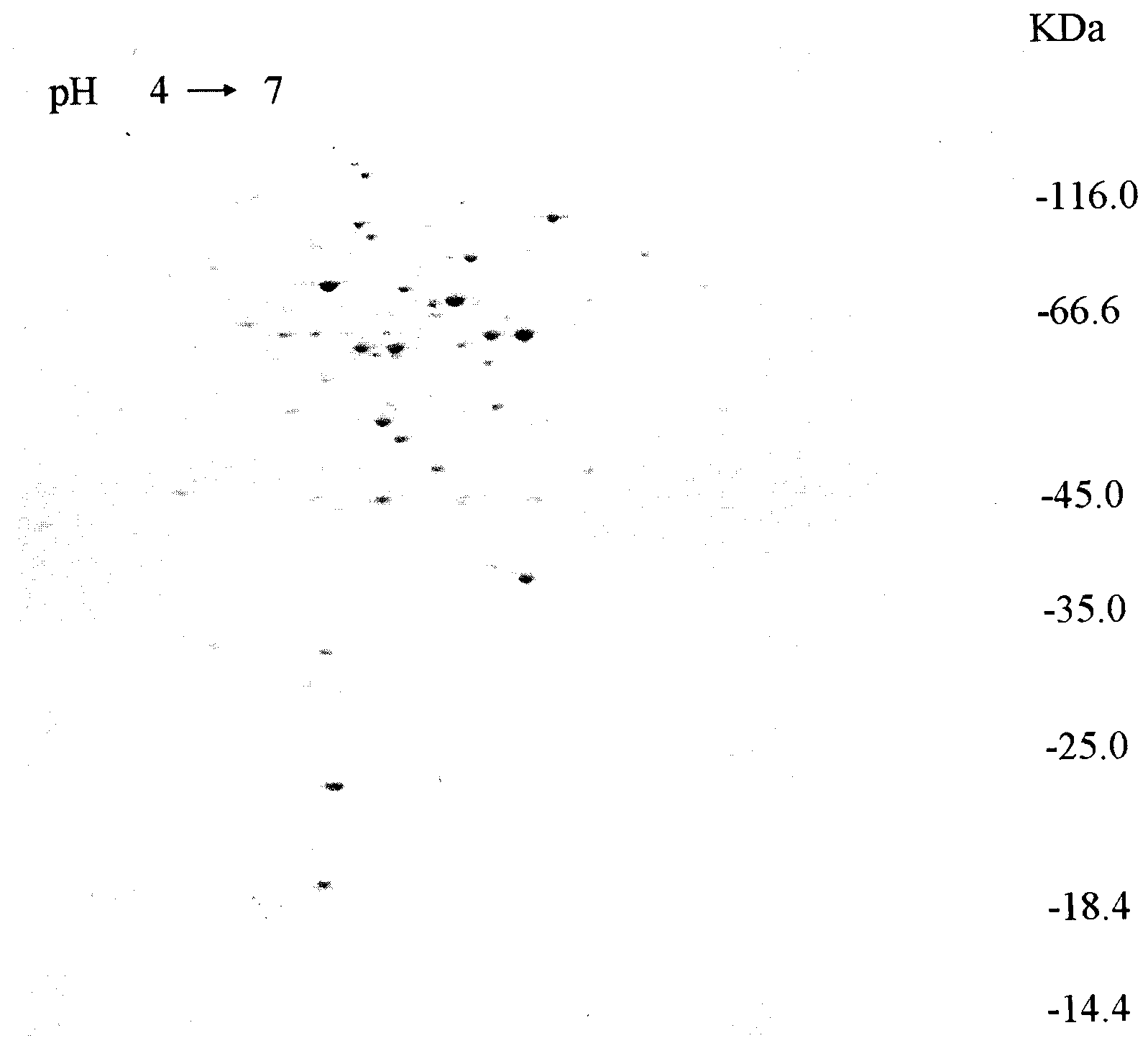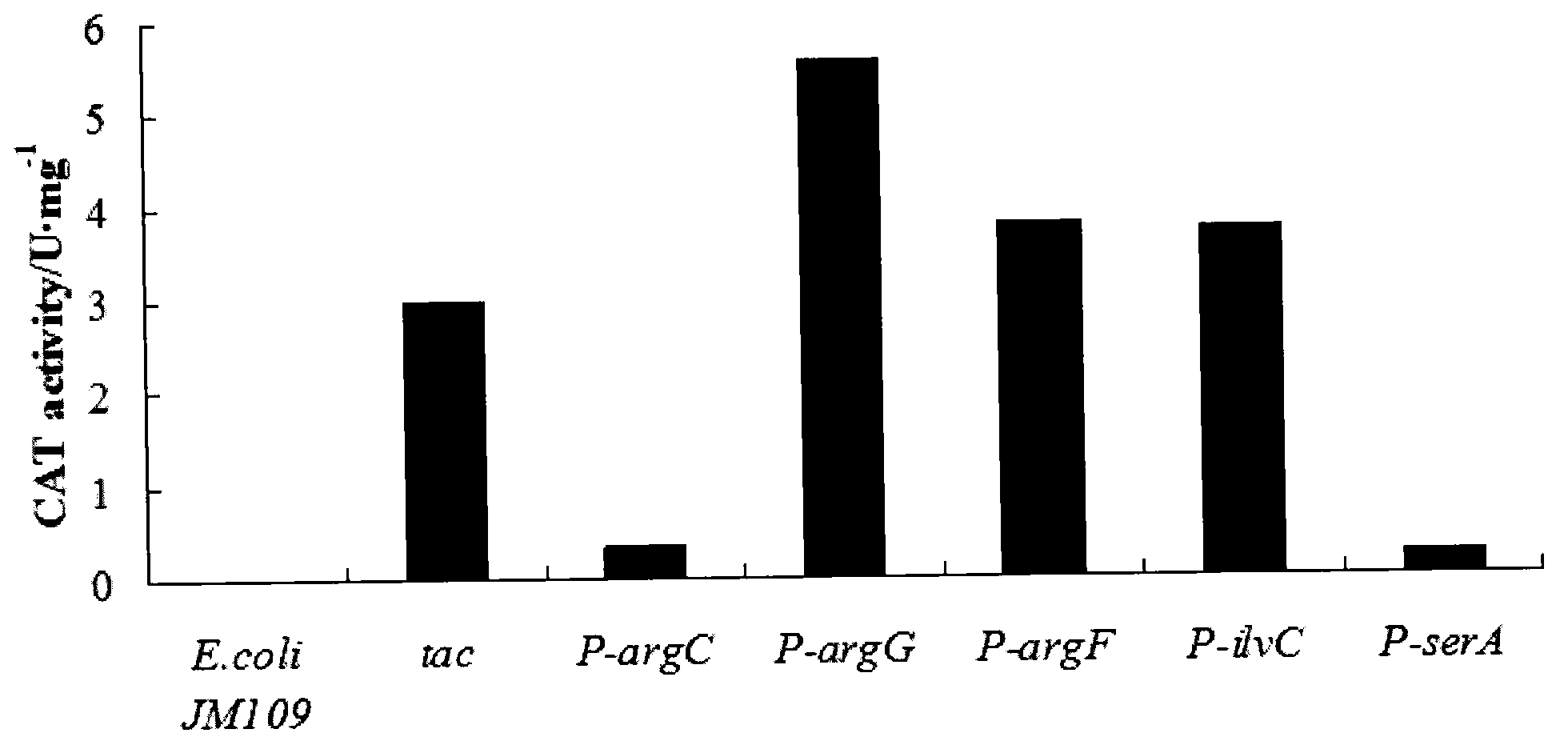Method for screening corynebacterium crenatum endogenous high-expression promoter by using 2-DE (Two-Dimensional Electrophoresis) technique
A Corynebacterium blunt tooth, 2-DE technology, applied in the field of proteomics and genetic engineering, can solve the problems of application limitations, low efficiency of exogenous promoters, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1: According to 2-DE screening corynebacterium bacillus endogenous highly active promoter
[0035] After culture and protein extraction, the extracted protein concentration is about 70 μg / μL. After the first dimension isoelectric focusing, equilibration and second dimension SDS-PAGE, after fixation, staining and decolorization, a condensate with clear protein distribution was obtained. Gel; use Lad Scan (GE Health) to scan the gel, and then use ImageMaster 2D Platinum5.0 software to adjust, find, quantify and match the gel image, and cut the selected protein spots for MALDI-TOF-MS And MS / MS analysis, a total of 15 highly expressed protein spots were successfully identified.
Embodiment 2
[0036] Embodiment 2: Construction of recombinant expression vector and bacterial strain
[0037] Based on the 15 highly expressed protein spots obtained in Example 1, combined with information such as their copy numbers in bacterial cells, 5 target protein spots were found, and the gene sequences corresponding to these 5 proteins were found in GENBANK , and separated its promoter sequence according to the gene sequence, and named these five promoters as P-argC, P-argG, P-argF, P-ilvC and P-serA. These five promoters were respectively replaced with the tac promoter on the shuttle vector pDXW-8, and the reporter gene chloramphenicol acetyltransferase gene cat gene was inserted downstream of it, and the recombinant vectors were transformed into Escherichia coli JM109 and Corynebacterium bacilli . After verification, the successful construction of pDXW-P argC -cat, pDXW-P argG -cat, pDXW-P argF -cat, pDXW-P ilvC -cat and pDXW-P serA -cat, and successfully obtained its corres...
Embodiment 3
[0038] Embodiment 3: the enzyme activity assay of recombinant bacterial strain
[0039] The recombinant strains were cultured in LB and LBG medium respectively, centrifuged at 8000rpm for 10min, washed twice with 100mM Tris-HCl buffer solution of pH 7.8, suspended in the buffer solution, and subjected to ultrasonic crushing to prepare crude enzyme solution. The reaction mixture is composed of a certain volume of crude enzyme solution, 100mM Tris-HCL (pH7.8), 0.1mM acetyl-CoA, and 0.4mg / mL DTNB, reacted at 37°C for 2min, added chloramphenicol to terminate the reaction, and detected Changes in absorbance at 412nm. The enzyme activity unit is defined as the amount of enzyme required to acetylate 1 μmol of chloramphenicol per minute.
[0040] The enzyme activity of the recombinant Escherichia coli was measured as image 3 Respectively: E.coli JM109 / pDXW-P argC -cat is 0.36U / mg, E.coli JM109 / pDXW-P argG -cat is 5.58U / mg, E.coli JM109 / pDXW-P argF -cat is 3.81U / mg, E.coli JM109 / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com