Mutant zymoprotein of D-amino acid oxidase and preparation method of mutant zymoprotein
An amino acid and oxidase technology, applied in the fields of genetic engineering and enzyme engineering, can solve the problems of high price and rising cost, and achieve the effect of improving activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 D-amino acid oxidase gene daao Acquisition and expression vector construction
[0061] (1) daao gene acquisition
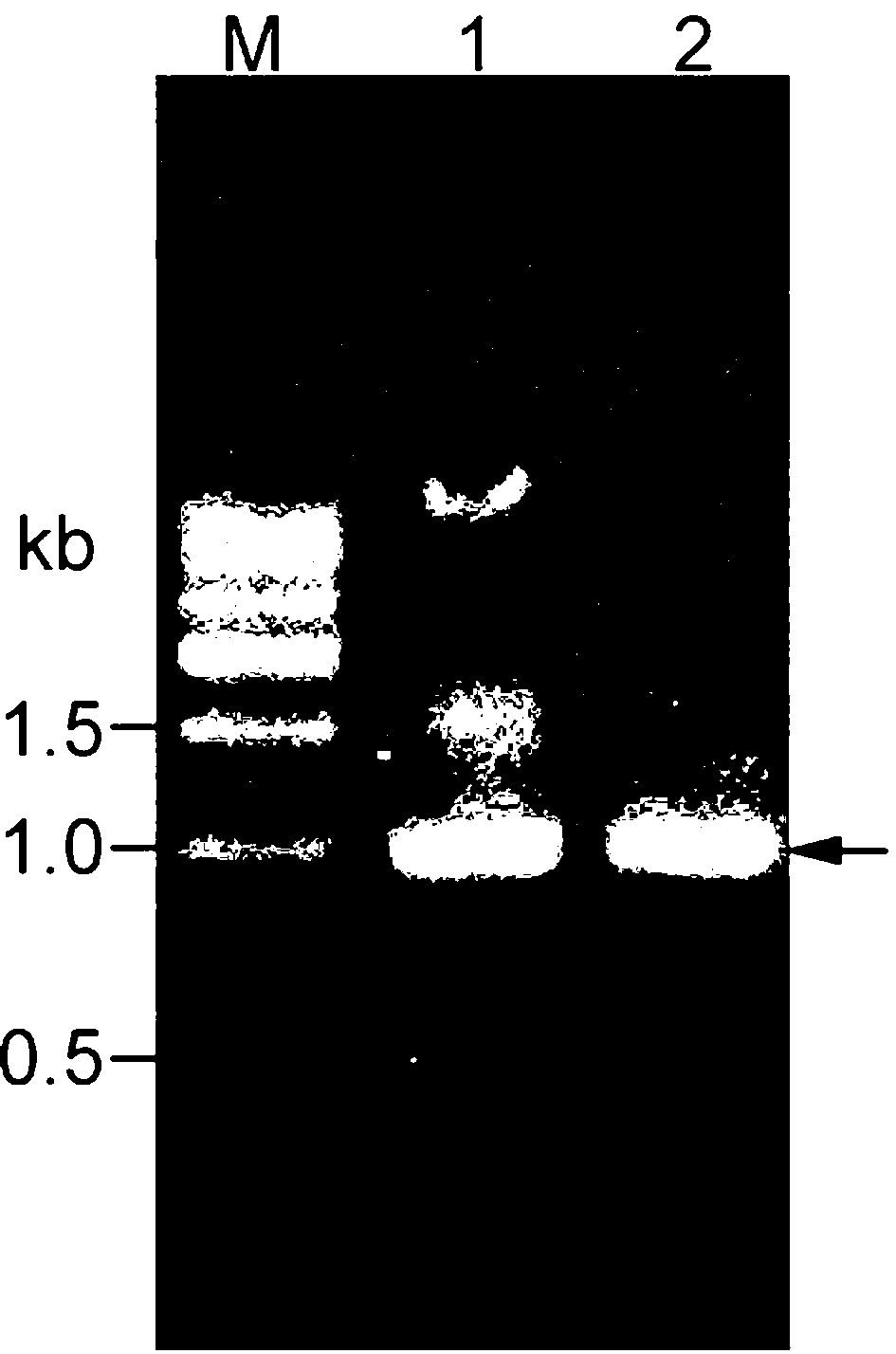
[0062] According to the original Arthrobacter glass fly ( Arthrobacter protophormiae , DSM20168) to design primers for the D-amino acid oxidase gene (GenBank ID. JX855922.1) to A. protophormiae Genomic DNA was used as a template, and a specific fragment was obtained by PCR amplification ( figure 1 , lane1). daao The amplification primers are:
[0063] Sense: 5′-TCC CATATG CCCACAGCACCGTTGAG-3′ (underlined as Nde I restriction site);
[0064] Anti-sense: 5′-ATA CTCGAG GCTGGCCGGCTCGCCA-3′ (underlined as xho I restriction site);
[0065] The PCR amplification reaction conditions were: pre-denaturation at 94°C for 4 min; denaturation at 94°C for 45 sec, annealing at 55°C for 1 min, extension at 72°C for 2 min, and 25 cycles; extension at 72°C for 10 min.
[0066] The amplified PCR fragment was recovered by gel, ligated with the vec...
Embodiment 2
[0070] Example 2 Mutant E115A / N119D / T286A expression vector construction
[0071] (1) Design mutation primers
[0072] According to the nucleotide sequences around 115, 119 and 286 of the D-amino acid oxidase to be mutated in the original Arthrobacter glass fly, design the following three pairs of site-directed mutagenesis primers:
[0073] The 115-position mutation primer is:
[0074] 115 anti-sense: 5'-CCTCCCGGGCGGATCTGC-3';
[0075] 115-bit sense: 5'-GGCAGATCCGCCCGGGAG-3';
[0076] The 119-position mutation primer is:
[0077] 119 anti-sense: 5′-TCTGCCGGACGGCGCCCAC-3′;
[0078] 119-position sense: 5′-TGGGCGCCGTCCGGCAGAT-3′;
[0079] The 286 mutation primers are:
[0080] 286 anti-sense: 5′-GAGCACGTCGCGGGCCAC-3′;
[0081] 286-bit sense: 5′-GTGGCCCGCGACGTGCTC-3′;
[0082] (2) Construction of mutant E115A expression vector
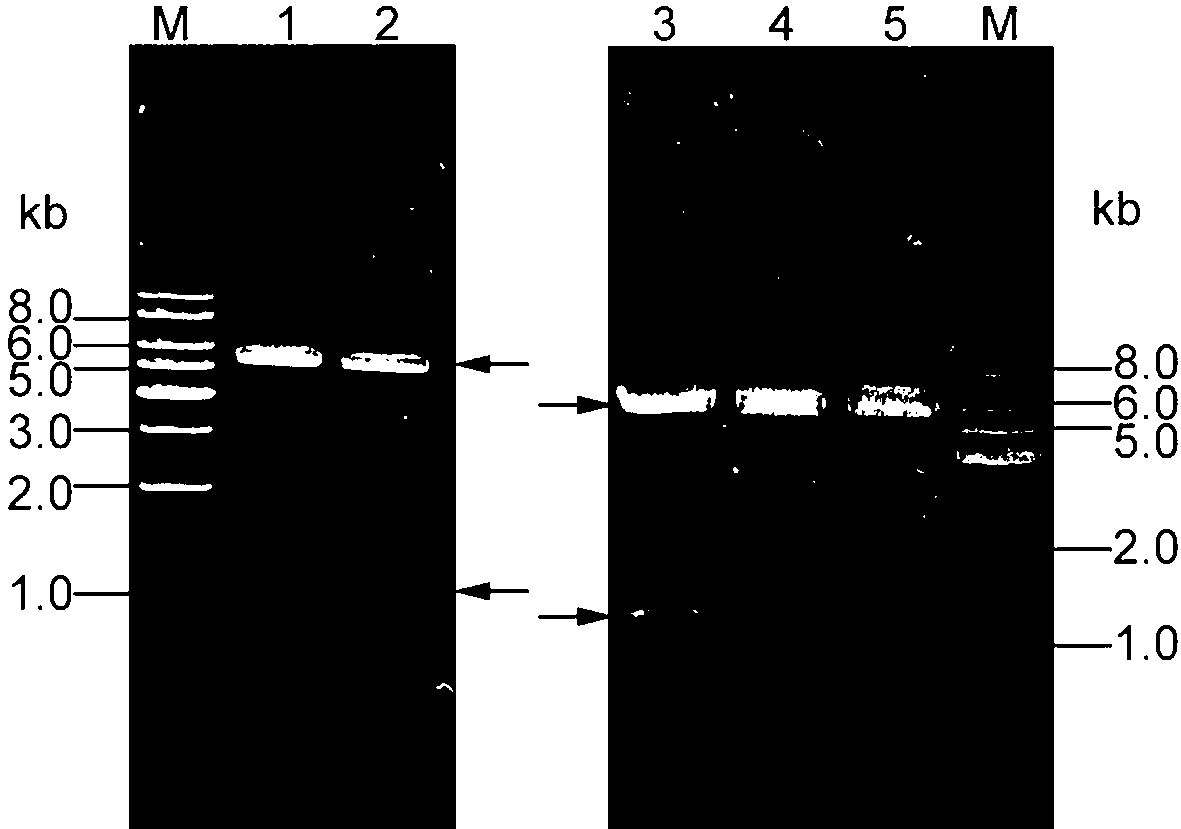
[0083] Based on the Quick Change Site-Directed Mutagenesis (QuickChange Site-Directed Mutagenesis), the recombinant expression vector pE...
Embodiment 3
[0089] Example 3 Expression and Purification of Mutant Enzyme and Wild-type Enzyme Protein
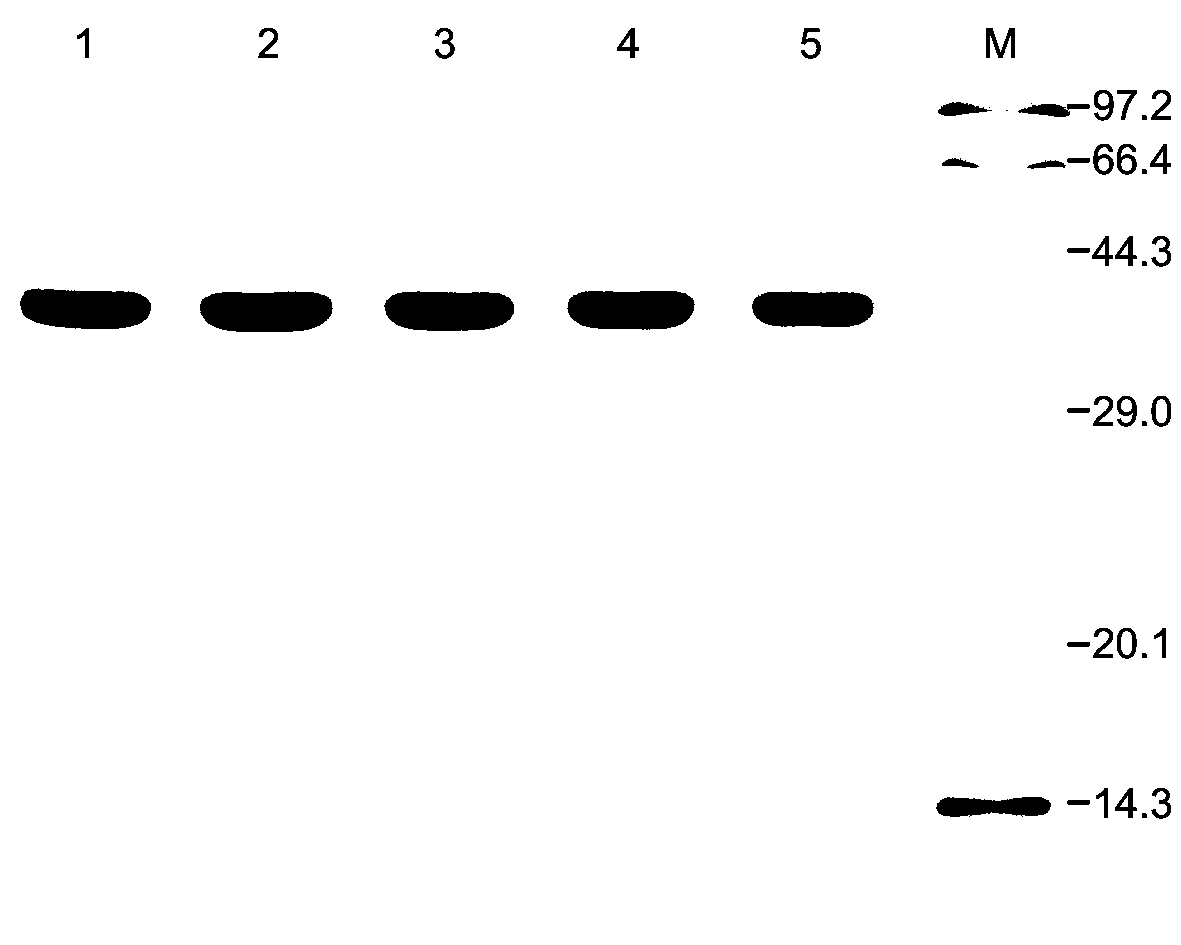
[0090] The plasmids pET-DAAO, pET-ApDAAO and the expression vectors of each mutant were transformed into Escherichia coli BL21(DE3) competent cells, and the transformants were selected and cultured overnight at 37°C in LB medium (containing 100 μg / mL ampicillin); Inoculate in 100mL LB liquid medium (containing 100μg / mL ampicillin) at a ratio of 1:100, culture at 37°C with shaking at 180rpm until OD 600 is 0.5, add 0.5mM isopropylthiogalactoside (isopropyl- β -D-thiogalactopyranoside, IPTG), induced expression overnight at 30°C. Centrifuge at 8000rpm for 10min at 4°C to collect the cells and ultrasonically disrupt them. Purify with Ni-NTA affinity chromatography, and the imidazole concentration in the eluent is 250mM to obtain purified wild-type proteins DAAO, ApDAAO, mutant proteins E115A, E115A / N119D and E115A / N119D / T286A.
[0091] The purified enzyme protein was identified by SD...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com