A method for preparing Escherichia coli that automatically degrades nucleic acid during lysis
A technology of automatic degradation of Escherichia coli, applied in the biological field, can solve the problems of high viscosity of lysate affecting purification, achieve the effect of reducing cytotoxicity, reducing the accumulation of acetic acid, and simplifying the purification steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
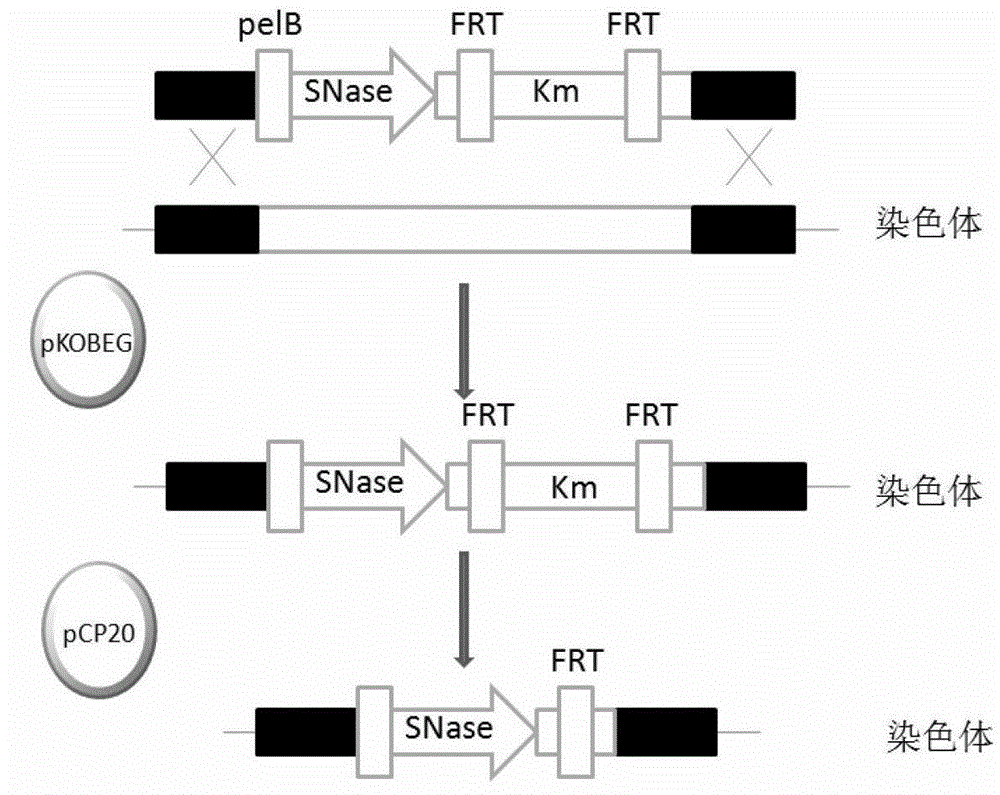
[0019] The embodiment of the present invention provides a method for preparing Escherichia coli that automatically degrades nucleic acid during lysis, comprising the following steps:
[0020] S01 connects the pelB signal peptide coding sequence with the Staphylococcus aureus nuclease gene sequence to obtain the first combination sequence, wherein the pelB signal peptide coding sequence is upstream of the Staphylococcus aureus nuclease gene sequence, and then the first combination The sequence is connected with the kanamycin resistance gene sequence to obtain a second combination sequence, wherein the first combination sequence is upstream of the kanamycin resistance gene sequence;
[0021] S02 replace the ptsG gene sequence of Escherichia coli JM109 with the second combined sequence prepared in step S01, to obtain Escherichia coli JM109 with ptsG gene deletion;
[0022] S03 Knockout the kanamycin resistance gene in the second combined sequence in Escherichia coli JM109 in whic...
Embodiment 1
[0055] 1. Obtaining SNase fragments
[0056] Synthesize forward and reverse primers according to the following sequence:
[0057] Forward primer P1: GGG GGATCC ATGAAGTCAAATAAATCGCTSEQ ID NO:3
[0058] Reverse primer P2: GGG CTCGAG TTATTTACTCCAAATATTTA SEQ ID NO: 4
[0059] Add the BamHI restriction site to the P1 primer, which is the underlined part, add the XhoI restriction site to the P2 primer, which is the underlined part, use P1 and P2 as primers, and use the Staphylococcus aureus genome as a template to prepare a 50 μL PCR reaction system, as follows:
[0060]
[0061] PCR reaction conditions:
[0062] Denaturation at 94°C for 5 minutes,
[0063] 94°C for 30s, 56°C for 1min, 72°C for 40s, 30 cycles,
[0064] Extend at 72°C for 10 min.
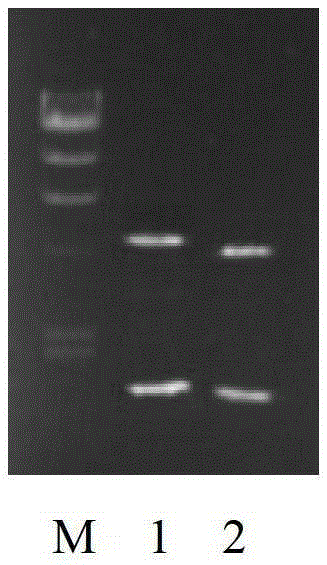
[0065] After the reaction, it was detected by electrophoresis on a 1% agarose gel containing 20 μg / mL ethidium bromide (EB). The length of the obtained target fragment was 552bp, which was verified by sequencing after PCR clon...
Embodiment 2
[0126] Steps 1-6 are the same as in Example 1.
[0127] 7. Transformation of pKD46 into Escherichia coli JM109 competent cells
[0128] Preparation of Escherichia coli JM109 competent cells.
[0129] The plasmid pKD46 was transformed into the Escherichia coli JM109 competent cells by heat shock method. Then pick a single clonal colony and culture overnight at 30°C with shaking in LB medium containing ampicillin (final concentration 34 μg / mL), then add 1 mL of the bacterial solution to 50 mL of LB medium with the same concentration of ampicillin, shake at 30°C Cultivate, to A 600 When it is 0.2-0.4, add L-arabinose (final concentration is 10mM), continue shaking culture, until A 600 centrifuge at 4000g for 10 minutes at 4°C, discard the supernatant, collect the bacteria, wash three times with deionized water, and finally resuspend the bacteria in 150 μL of pre-cooled 10% glycerol, and take 50 μL for electrotransformation.
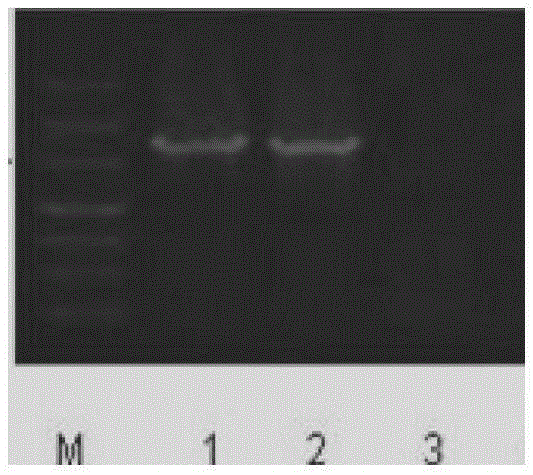
[0130] 8. Electrotransformation of the target gene...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com